Abstract
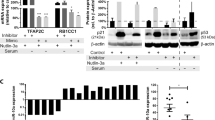
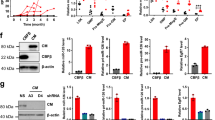
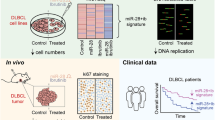
High levels of microRNA-155 (miR-155) are associated with poor outcome in acute myeloid leukemia (AML). In AML, miR-155 is regulated by NF-κB, the activity of which is, in part, controlled by the NEDD8-dependent ubiquitin ligases. We demonstrate that MLN4924, an inhibitor of NEDD8-activating enzyme presently being evaluated in clinical trials, decreases binding of NF-κB to the miR-155 promoter and downregulates miR-155 in AML cells. This results in the upregulation of the miR-155 targets SHIP1, an inhibitor of the PI3K/Akt pathway, and PU.1, a transcription factor important for myeloid differentiation, leading to monocytic differentiation and apoptosis. Consistent with these results, overexpression of miR-155 diminishes MLN4924-induced antileukemic effects. In vivo, MLN4924 reduces miR-155 expression and prolongs the survival of mice engrafted with leukemic cells. Our study demonstrates the potential of miR-155 as a novel therapeutic target in AML via pharmacologic interference with NF-κB-dependent regulatory mechanisms. We show the targeting of this oncogenic microRNA with MLN4924, a compound presently being evaluated in clinical trials in AML. As high miR-155 levels have been consistently associated with aggressive clinical phenotypes, our work opens new avenues for microRNA-targeting therapeutic approaches to leukemia and cancer patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Burnett A, Wetzler M, Löwenberg B . Therapeutic advances in acute myeloid leukemia. J Clin Oncol 2011; 29: 487–494.
Faraoni I, Antonetti FR, Cardone J, Bonmassar E . miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 2009; 1792: 497–505.
Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 2010; 285: 17869–17879.
Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrózek K et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol 2013; 31: 2086–2093.
Whitman SP, Maharry K, Radmacher MD, Becker H, Mrózek K, Margeson D et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood 2010; 116: 3622–3626.
O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 2008; 205: 585–594.
Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood 2009; 114: 1374–1382.
Georgantas RW, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA 2007; 104: 2750–2755.
O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D . Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA 2009; 106: 7113–7118.
Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007; 27: 847–859.
Gerloff D, Grundler R, Wurm AA, Bräuer-Hartmann D, Katzerke C, Hartmann JU et al. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia 2014; 29: 535–547.
Scott EW, Simon MC, Anastasi J, Singh H . Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 1994; 265: 1573–1577.
Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M . Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res 2008; 36: 6608–6619.
Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 2001; 98: 2301–2307.
Finco TS, Beg AA, Baldwin AS . Inducible phosphorylation of I kappa B alpha is not sufficient for its dissociation from NF-kappa B and is inhibited by protease inhibitors. Proc Natl Acad Sci USA 1994; 91: 11884–11888.
Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA . Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J 1995; 14: 2876–2883.
Chen ZJ, Parent L, Maniatis T . Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell 1996; 84: 853–862.
Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol 2000; 20: 2326–2333.
Petroski MD, Deshaies RJ . Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005; 6: 9–20.
Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB et al. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA 2000; 97: 4579–4584.
Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T . CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem 2008; 283: 29045–29052.
Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J 2006; 25: 1126–1136.
Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 2004; 24: 7130–7139.
Huh J, Piwnica-Worms H . CRL4(CDT2) targets CHK1 for PCNA-independent destruction. Mol Cell Biol 2013; 33: 213–226.
Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell 2010; 37: 102–111.
Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood 2010; 116: 1515–1523.
Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood 2010; 115: 3796–3800.
Swords RT, Savona MR, Maris MB, Erba HP, Hua Z, Faessel H et al. Pevonedistat (MLN4924), an Investigational, first-in-class NAE inhibitor in combination with azacitidine elderly patients with acute myeloid leukemia (AML) considered unfit for conventional chemotherapy: updated results from the phase I C15009 trial. Blood 2014; 124: abstract #2313.
Munker R, Nordberg ML, Veillon D, Williams BJ, Roggero A, Kern W et al. Characterization of a new myeloid leukemia cell line with normal cytogenetics (CG-SH). Leuk Res 2009; 33: 1405–1408.
Mueller BU, Pabst T, Hauser P, Gilliland G, Neuberg D, Tenen DG . Mutations of the transcription factor PU.1 are not associated with acute lymphoblastic leukaemia. Br J Cancer 2006; 94: 1918–1920.
Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA 2008; 105: 3945–3950.
Huang X, Schwind S, Yu B, Santhanam R, Wang H, Hoellerbauer P et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin Cancer Res 2013; 19: 2355–2367.
Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009; 458: 732–736.
DeAngelo DJ, Erba HP, Maris M, Swords RT, Anwer F, Altman JK et al. MLN4924, a novel investigational inhibitor of NEDD8-activating enzyme (NAE), in adult patients with acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS): results from multiple dosing schedules in a phase 1 study. Blood 2013; 122: abstract #1443.
Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA 2006; 103: 7024–7029.
Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood 2009; 114: 3265–3275.
Luo JM, Yoshida H, Komura S, Ohishi N, Pan L, Shigeno K et al. Possible dominant-negative mutation of the SHIP gene in acute myeloid leukemia. Leukemia 2003; 17: 1–8.
Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood 2011; 117: 4293–4303.
Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol 2001; 21: 3789–3806.
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129: 1401–1414.
Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA 2012; 109: E1695–E1704.
Ma X, Becker Buscaglia LE, Barker JR, Li Y . MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 2011; 3: 159–166.
Swords RT, Erba HP, DeAngelo DJ, Bixby DL, Altman JK, Maris M et al. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukemia and myelodysplastic syndromes: a phase I study. Br J Haematol 2015; 169: 534–543.
Metzner A, Precht C, Fehse B, Fiedler W . Stocking C, Günther A, et al. Reduced proliferation of CD34(+) cells from patients with acute myeloid leukemia after gene transfer of INPP5D. Gene Ther 2009; 16: 570–573.
Brauer H, Strauss J, Wegner W, Müller-Tidow C, Horstmann M, Jücker M . Leukemia-associated mutations in SHIP1 inhibit its enzymatic activity, interaction with the GM-CSF receptor and Grb2, and its ability to inactivate PI3K/AKT signaling. Cell Signal 2012; 24: 2095–2101.
Olweus J, Thompson PA, Lund-Johansen F . Granulocytic and monocytic differentiation of CD34hi cells is associated with distinct changes in the expression of the PU.1-regulated molecules, CD64 and macrophage colony-stimulating factor receptor. Blood 1996; 88: 3741–3754.
Zhang DE, Fujioka K, Hetherington CJ, Shapiro LH, Chen HM, Look AT et al. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol Cell Biol 1994; 14: 8085–8095.
Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG . CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol 1998; 18: 4301–4314.
Piloto O, Wright M, Brown P, Kim KT, Levis M, Small D . Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood 2007; 109: 1643–1652.
Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood 2006; 107: 293–300.
Moore AS . Faisal A, Gonzalez de Castro D, Bavetsias V, Sun C, Atrash B, et al. Selective FLT3 inhibition of FLT3-ITD+ acute myeloid leukaemia resulting in secondary D835Y mutation: a model for emerging clinical resistance patterns. Leukemia 2012; 26: 1462–1470.
Wang L, Zhang H, Rodriguez S, Cao L, Parish J, Mumaw C et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-κB-dependent manner. Cell Stem Cell 2014; 15: 51–65.
Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B . Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res 2011; 71: 3042–3051.
Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS et al. Treatment-emergent mutations in NAEβ confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell 2012; 21: 388–401.
Acknowledgements
We thank Millennium Pharmaceuticals Inc., Cambridge, MA, USA, for supplying MLN4924 compound and Donna Bucci for providing primary AML samples from the OSU Leukemia Tissue bank. We are thankful to Dr Massimo Mallardo (University of Naples, Naples, Italy) for making sequences of EMSA probes available. Drs Daniel Tenen and Annalisa DiRuscio kindly provided 503 and 503/PUER cell lines. This work was supported by National Science Foundation (EEC-0914790) for XH, LJL and RJL and the Wilmot cancer research fellowship for JHM. Grant Numbers: U10CA180861, R01 CA135332, P50 CA140158 for GM and MAC and the Gehr Family Center for Leukemia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Khalife, J., Radomska, H., Santhanam, R. et al. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia 29, 1981–1992 (2015). https://doi.org/10.1038/leu.2015.106
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.106
This article is cited by
-
Targeting NEDD8-activating enzyme for cancer therapy: developments, clinical trials, challenges and future research directions
Journal of Hematology & Oncology (2023)
-
Cytokine production in myelofibrosis exhibits differential responsiveness to JAK-STAT, MAP kinase, and NFκB signaling
Leukemia (2019)
-
miR-155 in cancer drug resistance and as target for miRNA-based therapeutics
Cancer and Metastasis Reviews (2018)
-
Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles
Scientific Reports (2017)
-
Synergistic anti-AML effects of the LSD1 inhibitor T-3775440 and the NEDD8-activating enzyme inhibitor pevonedistat via transdifferentiation and DNA rereplication
Oncogenesis (2017)