Abstract
Gliosarcoma (GS) is a rare variant (2%) of glioblastoma (GBM) that poses clinical genomic challenges because of its poor prognosis and limited genomic information. To gain a comprehensive view of the genomic alterations in GS and to understand the molecular etiology of GS, we applied whole-exome sequencing analyses for 28 GS cases (6 blood-matched fresh-frozen tissues for the discovery set, 22 formalin-fixed paraffin-embedded tissues for the validation set) and copy-number variation microarrays for 5 blood-matched fresh-frozen tissues. TP53 mutations were more prevalent in the GS cases (20/28, 70%) compared to the GBM cases (29/90, 32%), and the GS patients with TP53 mutations showed a significantly shorter survival (multivariate Cox analysis, hazard ratio=23.9, 95% confidence interval, 2.87–199.63, P=0.003). A pathway analysis showed recurrent alterations in MAPK signaling (EGFR, RASGRF2 and TP53), phosphatidylinositol/calcium signaling (CACNA1s, PLCs and ITPRs) and focal adhesion/tight junction (PTEN and PAK3) pathways. Genomic profiling of the matched recurrent GS cases detected the occurrence of TP53 mutations in two recurrent GS cases, which suggests that TP53 mutations play a role in treatment resistance. Functionally, we found that TP53 mutations are associated with the epithelial–mesenchymal transition (EMT) process of sarcomatous components of GS. We provide the first comprehensive genome-wide genetic alternation profiling of GS, which suggests novel prognostic subgroups in GS patients based on their TP53 mutation status and provides new insight in the pathogenesis and targeted treatment of GS.
Similar content being viewed by others
Introduction
Gliosarcoma (GS) is a rare type of brain tumor (annual incidence of 1 per 1 000 000), consisting of both malignant glial and mesenchymal components.1, 2, 3, 4 Although GS is currently treated in the same manner as glioblastoma (GBM), several lines of evidence suggest that GS is a distinct disease entity characterized by a specific histology, a worse prognosis and more frequent metastases to extracranial lesions compared to GBM,1, 3 which suggests that a different treatment approach for GS is needed.
The pathogenesis of GS is still largely unexplained. Early studies have suggested that the sarcomatous components originate from the mesenchymal components within the brain, such as endothelial cells.1, 2 However, subsequent studies have failed to detect endothelial markers in the sarcomatous cells of GS.5, 6, 7 An alternative hypothesis is a monoclonal origin for both the glial and sarcomatous cells. The sarcomatous cells are considered to arise through the mesenchymal differentiation of the malignant glial cells,1 which is also supported by similar genetic alterations in both the glial and mesenchymal components.4, 8, 9
Previous studies have reported genetic alterations of the target genes for a limited number of GS samples. PTEN mutations (15–45%),4, 9, 10, 11 TP53 mutations (24–73%)4, 9, 11 and TERT promoter mutations (83%)10 were common, and IDH1 mutations were rare or absent (0–7%).10, 11 At the chromosome level, the copy-number alterations (CNAs) that were commonly detected included amplifications of chromosome 7 (75%) and X (20%) as well as deletions of chromosome 10q (88%) and 9p (35%).4, 10 Amplifications of EGFR and homozygous deletions of CDKN2A were reported in 0–8%4, 9 and 37–60%9, 10 of the GS cases, respectively. However, a genome-wide characterization of the genetic alterations of GS is limited.
To provide a comprehensive view of the genomic alterations in GS, we generated an integrated genomic data set from whole-exome sequencing (WES) and a CNA analysis. Recurrent TP53 mutations were detected in 71% of the GS patients, and these patients showed a significantly shorter survival. In addition, the TP53 mutations were associated with the epithelial–mesenchymal transition process, suggesting that TP53 mutations have a role in GS development, treatment resistance and poor prognosis.
Materials and methods
Patient samples and genomic DNA extraction
Four normal blood/GS paired samples and two normal blood/GS/GBM trios were obtained for the discovery set. Twenty-three formalin-fixed paraffin-embedded GS samples, including two samples from one patient, were obtained for the validation set. The patients were diagnosed with GS through a standardized review by two independent expert pathologists at the Seoul National University Hospital and the Samsung Medical Center. Total DNA was extracted from the sections using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany). All samples were obtained with informed consent at the Seoul National University Hospital and the Samsung Medical Center, and the study was approved by the institutional review board in accordance with the Declaration of Helsinki.
Exome capture, library construction and sequencing
For all samples, 1 μg of DNA per sample was sheared using a Covaris SS Ultrasonicator (Covaris, Woburn, MA, USA). Exome capture was performed using an Agilent SureSelect Human All Exon Kit V5 (Agilent Technologies, Palo Alto, CA, USA). Each sample was sequenced on an Illumina HiSeq 2000 instrument using a read length of 2 × 101 bp. Image analysis and base calling were performed using the Illumina pipeline (Illumina, San Diego, CA, USA) with the default settings.
Exome sequencing processing and variant calling
All FASTQ files were aligned to the human reference genome GRCh37/hg19 by Burrows–Wheeler Aligner mem.12 Marking of the duplicate reads was performed by Picard tools. Indel realignment and base recalibration were performed using the Genome Analysis Tool kit.13 After processing the BAM files, MuTect was used to detect the somatic single nucleotide variants (SNVs), except for the TTN and MUC16 genes.14 ANNOVAR was used to add the annotations.15 To select functional variants, variants within the coding regions were retained and filtered by a minor allele frequency if <0.01 or not reported, based on the ExAC 65,000 exomes allele frequency data, the 1000 Genome Project for all population frequencies and the NHLBI Exome sequencing project 6500. Nonflagged SNPs, based on dbSNP138, were filtered out to sort out passenger calls, and possible false-positive calls in the segmental duplicate region were also removed. ADTEx and PatternCNV were used to detect somatic copy-number variations (CNVs) for the six blood/GS paired samples.16, 17
CNV microarray
The CNV microarray test was performed using the Affymetrix CytoScan HD array system according to the protocol provided by the manufacturer. The Affymetrix CytoScan HD array contains ~2.7 million probes throughout the human genome and includes 1 953 246 copy-number probes and 743 304 SNP probes. The current build used was the human genome reference build NCBI 37.3 (hg19). The build covers >36 000 RefSeq genes with 1 marker per 880 bases, complete International Standards for Cytogenomic Arrays (ISCA) constitutional coverage (1 marker per 384 bases), cancer gene coverage (1 marker per 553 bases), X chromosome genes (1 marker per 486 bases) and 12 000 OMIM genes (1 marker per 659 bases). Genomic imbalances were called using ChAS software (Affymetrix, Santa Clara, CA, USA) when a minimum of 25 consecutive probes were observed for a loss and when 50 consecutive probes were observed for a gain. To focus on the large chromosomal aberrations, a 2 Mb size cutoff is used for the CNV calls in this study.
Pathway analysis
Significantly altered pathways in GS were analyzed with a 201 gene list from the WES data of six normal/GS paired samples (discovery set) using the Kyoto Encyclopedia of Genes and Genomes analysis tool in the DAVID bioinformatics resource (http://david.ncifcrf.gov; P<0.1).18
Cell culture
U373-MG GBM cells were obtained from the Korean Cell Line Bank and were maintained in Dulbecco’s modified Eagle medium (Life Technologies, Gaithersburg, MD, USA) containing 10% fetal bovine serum (Life Technologies). The characteristics of the cells were tested and authenticated by the Korean Cell Line Bank, and cells in early passages were used for the experiment to minimize genetic changes. For the primary cell culture from the GS and GBM tissues, the samples were cut into pieces of ~1 mm on each edge and were incubated with trypsin-EDTA (33.3%; Life Technologies) and DNase I (20 U ml−1)-containing phosphate-buffered saline for 1.5 h at 37 °C. After an additional incubation with DNase I (20 U ml−1)-containing Dulbecco’s modified Eagle medium (10% fetal bovine serum included) for 15 min, the samples were triturated sequentially 20, 15 and 10 times through fire-polished glass pipettes with decreasing core diameters. After trituration, the samples were filtered through a 40-μm mesh, and the cells were eventually plated in T-75 culture flasks with Dulbecco’s modified Eagle medium containing 10% fetal bovine serum. Penicillin (100 U ml−1; Life Technologies) and streptomycin sulfate (100 μg ml−1; Life Technologies) were supplemented to all cell culture media. All cells were maintained in a humidified incubator with 5% CO2 at 37 °C.
Quantitative real-time PCR
Total RNA was purified using an RNeasy Plus Mini kit according to the manufacturer’s instructions (Qiagen). One microgram of total RNA was transcribed into complementary DNA using a Maxime RT PreMix (Intron Biotechnology, Seoul, Korea) for 1 h at 45 °C. Quantitative real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control for normalization. The sequences of the primers are summarized in Supplementary Table S1.
Gene set enrichment analysis
To identify the gene sets enriched in GS compared to GBM, we searched Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) for the gene expression profiling and selected the data set of GSE8692, contributed by Kenneth Kosik.19 From this data set, we chose a total of two GS samples and six GBM samples for further analysis. Each Affymetrix data set was background-adjusted and normalized by the Robust Multichip Averaging algorithm in the Affy package using R ver. 3.1.1.20 Gene set enrichment analysis (GSEA) was performed using the javaGSEA desktop application (GSEA v2.1.0).21, 22 For this analysis, the chemical and genetic perturbations (version 5.2) gene sets were used, and the gene sets with <50 genes and >100 genes were excluded. As the number of samples in this study was small, the P-values were calculated by permuting the data 1000 times for finding enriched gene sets. The GSEA software produces an enrichment score, a normalized enrichment score, nominal P-value and a false discovery rate (Q-value). The gene sets that were up- or downregulated with a Q-value<0.05 were considered significant. For the complementary DNA microarray data from the primary cell culture, the GSEA was performed with the same parameters except for using the pre-ranked options derived from the rank of fold changes of the GS samples compared to the GBM samples.
Complementary DNA microarray
Total RNA was extracted from the primary cells (<10 passages) using a TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The transcriptomic profiles of these glioma cells were characterized by an oligonucleotide microarray analysis using MA-human Agilent 44K (Agilent Technologies). The one-channel microarray data were analyzed by a global median normalization method using GeneSpring GX 7.3 (EBIOGEN, Seoul, Korea).
Site-directed mutagenesis and transfection of DNA
A QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) was used to generate point mutations in TP53, and the resulting mutations were verified by Sanger sequencing. The primers used in constructing the point mutations are listed in Supplementary Table S1. Transfection experiments were performed using Lipofectamine 2000 Transfection reagent according to the manufacturer’s protocol (Invitrogen). After 48 h, the cells were collected for real-time PCR and migration/invasion assays. The overexpression of the wild-type and mutants TP53 was validated by western blot analysis.
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Thermo Scientific, Hudson, NH, USA) containing a protease inhibitor cocktail (Roche, Indianapolis, IN, USA) and a phosphatase inhibitor cocktail (Roche) and were centrifuged at 20 000 g for 10 min at 4°C. After determining of the protein concentration in the cell extract by the bicinchoninic acid (BCA) method (Thermo Scientific), 20 μg of protein was resolved by SDS-polyacrylamide gel electrophoresis and was transferred to a polyvinyl difluoride membrane. Membranes were blocked for 1 h with 5% skim milk in Tris-buffered saline and were incubated with anti-Flag (Sigma-Aldrich Corporation, St Louis, MO, USA) and anti-actin (Sigma-Aldrich Corporation) antibodies. The membranes were washed and were incubated with a horseradish peroxidase-conjugated secondary antibody, followed by enhanced chemiluminescence development according to the manufacturer’s instructions (Pierce, Rockford, IL, USA).
In vitro migration and invasion assay
Transfilter migration and invasion assays were performed using 8.0-μm pore inserts in a 24-well Transwell (BD Biosciences, San Jose, CA, USA). Cells were transfected with wild-type and mutant TP53-expressing vectors using Lipofectamine 2000 Transfection reagent. After 24 h of incubation, the cells were isolated and added to the upper chamber of a transwell (5 × 103 cells per well) with a non-coated filter and were incubated for 24 h in the migration assay. The invasion assays were performed using 12.5% Matrigel (Corning, New York, NY, USA)-coated filters at 5 × 103 cells per well, and the cells were incubated for 48 h. The migrated or invaded cells were stained with a Diff-Quik staining kit (Sysmex Corporation, Kobe, Japan). Each experiment was performed in triplicate.
Statistical analysis
Statistical calculations were performed using SPSS, version 22.0 (IBM Corporation, Somers, NY, USA) or Prism 4.0 (GraphPad, San Diego, CA, USA). Kaplan–Meier curves were constructed to evaluate overall survival. For univariate survival analysis, log-rank tests were used to explore the differences in survival according to the TP53 mutation status. Multivariate Cox analyses were performed to evaluate the predictive factors of overall survival. The variables tested included sex (categorical), age (continuous), tumor size (continuous), tumor location (categorical), treatment (categorical) and TP53 mutation status (categorical). The differences between multiple variables were assessed by a one-way analysis of variance with a Tukey’s multiple comparison test. The difference was considered significant if the P-value was <0.05.
Results
Genome-wide analyses of mutations and CNAs in GS
We explored genomic alterations in GS through WES of six paired tumor and normal samples (the discovery set). In addition, we analyzed additional 22 formalin-fixed and paraffin-embedded GS samples from a multicenter study (the validation set). Detailed clinical information and the magnetic resonance imaging images for these patients are provided in Table 1, Supplementary Table S2 and Supplementary Figure S1. On average, 99.9% of the reads mapped for all samples, with an average read coverage of 124.64 × and 117.95 × for the six matched normal samples and the six GS samples, respectively. Similarly, the average read coverage for the 22 formalin-fixed paraffin-embedded samples was 126.76 ×.
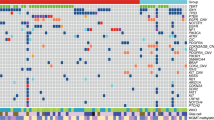
In the discovery set, exome sequencing identified a total of 295 somatic mutations (199 missense mutations, 9 nonsense mutations, 77 synonymous mutations, 5 splice sites mutations and 5 unclassified mutations). The 213 non-silent mutations were matched to 201 genes (Supplementary Table S3). Somatic mutations in TP53 were the most frequent alterations (5/6), and mutations of PTEN, SETD2, PLCL1, BCAM and PCDH19 were also recurrently detected (Figure 1a; Supplementary Table S3).
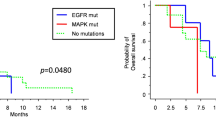
Genomic alterations of gliosarcoma (GS) patients. (a) Mutational profiles for 28 GS patients by whole-exome sequencing. Matrix of the altered genes colored by types of mutations. Each column stands for an individual cancer patient (#1–6 for discovery set and #7–28 for validation set), and each row denotes a gene. The right bar graph represents the number of samples with mutations for each gene in the 28 GS patients. (b) Left panel: Kaplan–Meier plot for the overall survival of the GS patients with (n=20; green line) and without TP53 mutations (n=8; blue line). Right panel: Kaplan–Meier plot for the overall survival in the Korean glioblastoma (GBM) patients with (n=30; green line) and without TP53 mutations (n=61; blue line). (c) Copy-number alterations for chromosome 7 (Chr7) and chromosome 10 (chr 10) in five GS patients. The amplifications (blue) and deletions (red), determined by a copy-number variation (CNV) microarray, are displayed for chromosomes 7 and 10.
In the 22 patient validation set, 5904 somatic or germline mutations (5619 nonsynonymous mutations, 185 nonsense mutations, 5 stoploss mutations and 95 splice sites mutations; Supplementary Table S4) were identified. To preclude the possible germline mutations, we further focused on the 201 genes detected to have non-silent mutations in the discovery set. The TP53 mutation was also found as the predominant mutation type in the validation set (15/22, 68.2%), and all TP53 mutations, except for one, were already reported in the COSMIC database (Supplementary Table S5). Therefore, alterations in TP53 were detected in 71% of all GS cases (20/28; Figure 1a; Supplementary Table S6), which were significantly higher than in cases of Korean GBM patients (29/90, 32%, Supplementary Table S7). The mutation abundance was not significantly different between the TP53 wild-type and the TP53-mutated groups (P=0.3884). In addition, we identified 43 recurrently mutated genes in three or more cases from the total 28 GS samples. PTEN mutations were the second most frequent alteration observed (8/28, 28.6%; Figure 1a; Supplementary Table S6).
Clinically, TP53-mutated GS appeared to have a poorer prognosis than non-mutated TP53 GS in both a univariate analysis (log-rank test, P=0.019; Figure 1b) and the multivariate analyses (multivariate Cox analysis, hazard ratio=23.9, 95% confidence interval, 2.87–199.63, P=0.003; Table 2), which suggest that TP53-mutated GS is a unique subgroup of GS. Other clinical parameters, such as location, size, gender and GS type (primary or secondary), showed no association between the TP53 wild-type and the TP53-mutated subgroups (Supplementary Table S8). By contrast, no significant survival difference was observed between the TP53 wild-type and the TP53-mutated subgroups in the Korean GBM patient data (n=90, log-rank test, P=0.368; Figure 1b, detailed clinical information is in Supplementary Table S7). Patients with mutations in PTEN from our GS group showed little difference in overall survival compared to those without mutations (log-rank test, P=0.819, Supplementary Figure S2).
A gain of chromosome (chr) 7 and a loss of chr 10 are well-known events that occur during the evolution of cells into GBM.23 For the somatic CNA analysis, we applied the genome-wide CNV microarray platform for the five paired tumor and normal samples (Supplementary Figure S3; Supplementary Table S9). Chr 7 was amplified in all the GS samples, and chr 10p was deleted in two out of the five cases analyzed from the discovery set (Figure 1c). These alterations were consistent with the copy-number analysis results using the WES data (Supplementary Figure S4). The copy-number analysis using WES also showed well-known CNAs, such as amplification events of EGFR, CDK6 and MET, as well as heterozygous deletions of PTEN, RB1 and NF1 in the GS samples from the discovery set (Supplementary Table S10). These findings imply that GS has some shared alterations with GBM in terms of CNA.
A biallelic loss of TP53 probably has profound effects on cancer cell development. Out of the five patients (patient #1–5) whose WES and CNV microarray data were available, we found one patient (patient #2) showing biallelic alterations of TP53 with both mutation (D149G) from WES data and copy-number loss from the CNV microarray data (Supplementary Figure S5; Supplementary Table S11).
Significantly altered pathways in GS
We performed pathway analysis on the 201 altered genes identified in the discovery set using the Kyoto Encyclopedia of Genes and Genomes analysis tool24 in the DAVID bioinformatics resource (http://david.ncifcrf.gov).25 Several important pathways, such as MAPK signaling, phosphatidylinositol/calcium signaling and focal adhesion/tight junction pathways, were significantly altered in the GS cases (Figure 2a and 2b,; Supplementary Tables S12 and S13). Mutations in genes of the MAPK signaling pathway were detected in 79% (22/28) of the GS cases, which included EGFR, FGFR1, RASGRF2 and TP53. We also found mutations in genes of the phosphatidylinositol/calcium signaling pathway, which included CACNA1F/1I, PLCB3/L1 and ITPR1/3, in 64% (18/28) of the GS cases. Finally, we found mutations in the genes of the focal adhesion/tight junction pathway, which included COL5A1, ITGB7, PAK3 and PTEN, in 57% (16/28) of the GS cases (Figure 2a and 2b,; Supplementary Table S13).
Pathway analysis in gliosarcoma (GS). (a) List of the somatically mutated pathways analyzed by the Kyoto Encyclopedia of Genes and Genomes database (P<0.1). The left side shows the number of affected patients out of the 28 patients in each pathway. The middle matrix indicates the alteration events in each individual (column), and the patients were clustered according to altered pathways by a hierarchical cluster analysis. The right side shows the P-values from the pathway analysis using DAVID bioinformatics resource. (b) Mutated genes in the MAPK signaling, phosphatidylinositol/calcium signaling and focal adhesion/tight junction pathways. Mutation frequencies are expressed as a percentage of cases (n=28).
Analysis of genomic alteration profiles in the matched initial and recurrent GS samples
Among our GS patient cohort, we found three patients with matched initial and recurrent tumors after treatment (surgery and concurrent chemoradiotherapy with temozolomide), and then we compared the mutational profiles between the initial and recurrent samples. Patient #5 was first diagnosed as having GS through a pathological examination; after treatment, the tumor recurred as a same-site GBM at the second operation and a same-site GS at the third operation (Figure 3a and 3b,). We detected 56 and 21 recurrence-specific mutations in first recurrent sample (GBM) and second recurrent sample (recurrent GS), respectively (Figure 3b; Supplementary Table S14). Interestingly, the WES analysis detected a TP53 mutation only in the recurrent GS sample (Figure 3b; Supplementary Table S14). A comparison of the CNAs between the initial GS and the recurrent GBM in patient #5 showed increased CNAs in the recurrent sample, especially in chromosomes 4, 5 and 21 (Supplementary Figure S6), suggesting that CNAs are also associated with treatment resistance.
Changes in histology, magnetic resonance imaging (MRI) images and the mutational profile in recurrent gliosarcoma (GS) patients. (a) Representative microscopic images of the GS and glioblastoma (GBM) tissues of patient #5, who had same-site recurrent samples showing the conversion of pathological diagnosis (initial GS to the same-site recurrent GBM after the first treatment and GBM to the same-site recurrent GS after the second treatment). The gliomatous and sarcomatous areas were visualized by H&E staining (× 200 in initial GS, and × 100 in GBM and recurrent GS). Gliomatous area was positive for glial fibrillary acidic protein (GFAP) staining (× 40 in initial GS, and × 100 in GBM and recurrent GS), and sarcomatous area showed Masson trichrome (MT)-positive collagen deposit (200 ×). Scale bar, 100 μm. (b) MRI images (left panel) and Venn diagrams representing the number of mutated genes (right panel) in three patients (patients #5, #6 and #22) who had matched initial and recurrent tumors after treatment. The genes in the Venn diagram are the mutated genes selected from the modified JAX Cancer Treatment Profile.44 Asterisks indicate mutations detected by droplet digital PCR. WT, wild type.
Patient #6 was first diagnosed as having GBM; however, after treatment, the tumor recurred as a same-site GS (Figure 3b). We found only one recurrence-specific mutation (ZFN469) in patient #6, and the TP53 mutation was detected in both the initial and recurrent GS samples (Figure 3b; Supplementary Table S15). To preclude the possibility of false positives in the WES, we performed Sanger sequencing for mutations found in one of the matched samples, and four initial tumor-specific mutations (HEPACAM2, CCDC83, PTPRT and FTSJ1) and one recurrence-specific mutation (ZNF469) were validated (Supplementary Table S15).
An additional recurrent sample from patient #22 was determined as GS in the pathological investigation of both the initial and same-site recurrent lesions. Of the 201 genes found in the discovery set, the WES analysis detected three mutations (ITPR3, GIMAP8 and TP53) only in the recurrent sample, likely allowing these clones to overcome therapy (Figure 3b; Supplementary Table S16). Interestingly, TP53 mutations were found in all three recurrent GS samples, and the WES analysis detected TP53 mutations in the recurrent GS samples from patient #5 and #22 (Figure 3b). Moreover, the amino-acid changes of these two patients were in the same residue (D281G; Figure 3b). Therefore, TP53 mutations are likely associated with the outgrowth of aggressive GS subclones that are resistant to previous treatments, such as concurrent chemoradiotherapy with temozolomide.
To investigate whether the TP53 mutations in the recurrent tumors existed as a clone in the initial tumors, we adopted droplet digital PCR, which is a highly sensitive method to detect mutations to a 0.1% frequency within a sample.26 We detected a D281G TP53 mutant allele at a frequency of 0.24% and 0.12%, in the initial GS and the same-site recurrent GBM samples of patient #5, respectively (Supplementary Figure S7a). Similarly, we found a D281G TP53 mutation at a frequency of 1.7% in the initial GS of patient #22 (Supplementary Figure S7b). After treatment of the initial GS of patient #22, we detected the TP53 mutations at a frequency of 82.2% in the recurrent sample (Supplementary Figure S7b). These results suggest that TP53 mutations exist as a minor clone at a low allele frequency in the initial tumors, and that TP53 mutations may play a role in treatment resistance.
Association of TP53 mutations with the EMT process
Given that EMT is the conversion of cells with epithelial characteristics into cells with mesenchymal properties,27 we investigated the role of the EMT process in GS development. Concordantly, the EMT signature gene expressions, including N-cadherin, vimentin and α-smooth muscle actin,28 were elevated in the GS tissues of patients #5 and #6 compared to the GBM tissues (Figure 4a). In addition, the GSEA using the microarray data from the Gene Expression Omnibus database (GSE8692) showed that the two cancer EMT-associated gene sets (ANASTASSIOU_CANCER_MESENCHYMAL_TRANSITION_SIGNATURE29 and JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP30) were significantly enriched in GS compared to GBM (Supplementary Figure S8; Supplementary Table S17). In addition, the GSEA analysis showed that inflammation-related signatures, including HECKER_IFNB1_TARGETS, SANA_RESPONSE_TO_IFNG_UP, SANA_TNF_SIGNALING_UP, MORI_MATURE_B_ LYMPHOCYTE_UP, LU_IL4_SIGNALING and COATES_MACROPHAGE_M1_VS_ M2_UP, were enriched in the GS samples compared to the GBM samples. An enrichment of the cancer EMT-associated gene sets in the GS samples was also reproduced in the GSEA analysis using a complementary DNA microarray data from the primary culture cells of the GS and GBM tissue from patient #6 (Supplementary Table S18), suggesting that EMT processes are involved in the mesenchymal differentiation in GS.
Association of the epithelial–mesenchymal transition signature with TP53 mutations. (a) Relative mRNA expression levels of EMT-associated genes in gliosarcoma (GS) and glioblastoma (GBM) tissues from patients #5 and #6. The amount of mRNA for each gene was determined by real-time PCR. (b) The locations of somatic mutations of TP53 in the 28 GS samples (red circles). The protein of TP53 consists of a transactivation domain (TAD), an SH3 domain, a DNA-binding domain (DBD), a tetramerization domain (TD) and a regulation domain (RD). Asterisks indicate recurrently mutated residues in the TP53 gene. (c) Regulation of EMT-associated genes by wild-type and mutant TP53 in the mutant TP53 (R273H)-expressing cells (U373-MG). The effects of wild-type and mutant TP53 on mRNA expression of EMT-associated genes were estimated by real-time PCR. (d) Effect of the wild-type and mutant TP53 on cell migration and invasion in the mutant TP53 (R273H)-expressing cells (U373-MG). In vitro cell migration and invasion were estimated by a transfilter migration and invasion assay. Left graphs show the number of migrating (upper) and invading (lower) cells from triplicate experiments. The right panel is the representative images of the migrating (upper) and invading (lower) cells. Asterisks indicate significant differences (*P<0.05; **P<0.01; ***P<0.001) compared to wild-type TP53.
We found 18 mutated residues of the TP53 gene in 20 GS patients, and four mutations (R158H, R248W, D281G and R342X) were recurrently detected in two or more cases of GS (Figure 4b; Supplementary Table S5). All TP53 mutations, except one stopgain mutation (R342X), were detected in the DNA-binding domain of TP53 (Figure 4b). To investigate the role of TP53 mutations in the EMT signature of GS, we examined the effects of the TP53 mutants on EMT-associated gene expression. The overexpression of wild-type TP53 in the mutant TP53-expressing cells (U373-MG) attenuated the expression of N-cadherin and vimentin (Figure 4c). Among the randomly selected six TP53 mutants (five mutants from DNA-binding domain and the one mutant from tetramerization domain), five mutants (R158H, R175H, P190S, D281G and R342X) showed a decreased inhibitory effect on the mRNA expression of either or both N-cadherin and vimentin compared to wild-type TP53 (Figure 4c). In addition, these mutants have functional defects in inhibiting cell viability compared to the wild-type TP53 in clonogenic assays (Supplementary Figure S9).
EMT increases the migration and invasion of cancer cells, resulting in increased metastasis.31 The overexpression of wild-type TP53 decreased in vitro migration and invasion of the mutant TP53-expressing cells (U373-MG; Figure 4d). Out of the randomly selected six TP53 mutants, the R158H and P190S mutants had reduced inhibitory effects on cell migration and invasion compared to the wild-type TP53 (Figure 4d), which are characteristics of EMT. Therefore, the high prevalence of TP53 mutations in GS is associated with EMT features.
Discussion
In this study, we found dominant mutations of the TP53 gene (20/28, 71%) in GS, and somatic mutations of the TP53 gene are associated with a poor patient survival, suggesting that the GS patients can be divided into two main molecular subgroups according to their TP53 mutation status. By comparing the paired initial/recurrent samples from the same patient, we found two cases in which the TP53 mutations were detected only in the WES analysis of the recurrent samples, which are likely associated with treatment resistance. In addition, we showed that the TP53 mutations rendered the cells more vulnerable to mesenchymal differentiation via cancer EMT-like processes (Supplementary Figure S10).
GS is considered a rare variant of GBM,1 and extensive studies of the genomic alterations in GBM have been performed.32, 33 Mutations in IDH1 were found to be a molecular marker that distinguished primary and secondary GBM.34 The most frequent genetic alteration of GBM (~60%) is a deletion at 10q,35, 36 and TP53 mutations are common in secondary GBM (~25%) but are rare (<15%) in primary GBM.37 Using the expression profiles of GBM, proneural, neural, classical, mesenchymal and proliferative patterns were reported.33, 38 Most IDH1-mutated GBM (92%) show a proneural expression signature, and high frequencies of TP53 (54%) and NF1 (37%) mutations are characteristic of the proneural and mesenchymal subtypes, respectively.33 We found one patient with an IDH1 mutation and another patient with an NF1 mutation out of the 28 GS patients (Supplementary Table S6) and detected a hemizygous deletion of NF1 in three of the GS samples within the discovery set (Supplementary Table S10). Therefore, GS showed distinctive genomic alteration profiles in the known cancer-driver genes compared to the subtypes of GBM.
EMT is a process by which epithelial cells undergo changes into mesenchymal cells, which are usually associated with cancer aggressiveness.27, 39 EMT is often induced by epigenetic reprogramming mediated by EMT-inducing transcription factors, such as Twist, Snail, Slug and Zeb1.39 A previous study reported significant nuclear localization of Twist and Slug in the mesenchymal tumor area of GS samples, suggesting that GS is associated with EMT.40 Notably, the MAPK signaling, phosphatidylinositol/calcium signaling and focal adhesion/tight junction pathways, which are significantly altered in GS cases (Figure 2a), are known to cooperate in the initiation and progression of EMT.41, 42 In addition, losses or mutations in the TP53 gene were previously shown to correlate with the activation of EMT reprogramming as well as increased stem cell properties by modulating microRNAs.43 Therefore, TP53 mutations induce the epigenetic reprograming of EMT genes, which could be the molecular mechanism of the mesenchymal differentiation, increased aggressiveness and poor prognosis of GS.
In addition to the roles of TP53 mutations in the development of GS via mesenchymal differentiation, our data suggested that TP53 mutations were also associated with the treatment resistance of GS. We found two patients (#5 and #22) in whom TP53 mutations were only found in the recurrent GS samples by the WES analysis. However, the more sensitive droplet digital PCR method detected TP53 mutations in the preceding lesions, although the allele frequencies were very low. In these patients, most of the cells in preceding GS regions did not have TP53 mutations, suggesting that the development of the initial GS was not highly associated with TP53 mutations. However, minor clones with TP53 mutations survived after treatment and became one of major populations in the recurrent GS sample, suggesting that TP53 mutations were associated with treatment resistance.
In summary, our study provides the first understanding of the genomic alterations specific to GS and elucidates the relationship among predominant mutations of TP53, EMT process and poor prognosis in GS. These results shed light on the understanding of the pathogenesis and clinical characteristics of GS for future therapeutic applications.
Data deposition
The sequence reported in this paper has been deposited in the European Nucleotide Archive (accession no. PRJEB12063).
References
Han SJ, Yang I, Tihan T, Prados MD, Parsa AT . Primary gliosarcoma: key clinical and pathologic distinctions from glioblastoma with implications as a unique oncologic entity. J Neurooncol 2010; 96: 313–320.
Feigin IH, Gross SW . Sarcoma arising in glioblastoma of the brain. Am J Pathol 1955; 31: 633–653.
Kozak KR, Mahadevan A, Moody JS . Adult gliosarcoma: epidemiology, natural history, and factors associated with outcome. Neuro Oncol 2009; 11: 183–191.
Actor B, Cobbers JMJL, Buschges R, Wolter M, Knobbe CB, Lichter P et al. Comprehensive analysis of genomic alterations in gliosarcoma and its two tissue components. Genes Chromosomes Cancer 2002; 34: 416–427.
Grant JW, Steart PV, Aguzzi A, Jones DB, Gallagher PJ . Gliosarcoma: an immunohistochemical study. Acta Neuropathol 1989; 79: 305–309.
Perry JR, Ang LC, Bilbao JM, Muller PJ . Clinicopathologic features of primary and postirradiation cerebral gliosarcoma. Cancer 1995; 75: 2910–2918.
Sreenan JJ, Prayson RA . Gliosarcoma. A study of 13 tumors, including p53 and CD34 immunohistochemistry. Arch Pathol Lab Med 1997; 121: 129–133.
Biernat W, Aguzzi A, Sure U, Grant JW, Kleihues P, Hegi ME . Identical mutations of the p53 tumor suppressor gene in the gliomatous and the sarcomatous components of gliosarcomas suggest a common origin from glial cells. J Neuropathol Exp Neurol 1995; 54: 651–656.
Reis RM, Konu-Lebleblicioglu D, Lopes JM, Kleihues P, Ohgaki H . Genetic profile of gliosarcomas. Am J Pathol 2000; 156: 425–432.
Oh JE, Ohta T, Nonoguchi N, Satomi K, Capper D, Pierscianek D et al. Genetic alterations in gliosarcoma and giant cell glioblastoma. Brain Pathol 2015; 125: 401–410.
Cachia D, Kamiya-Matsuoka C, Mandel JJ, Olar A, Cykowski MD, Armstrong TS et al. Primary and secondary gliosarcomas: clinical, molecular and survival characteristics. J Neurooncol 2015; 125: 401–410.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013; 11: 11.10.1–11.10.33.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219.
Wang K, Li M, Hakonarson H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164.
Amarasinghe KC, Li J, Hunter SM, Ryland GL, Cowin PA, Campbell IG et al. Inferring copy number and genotype in tumour exome data. BMC Genomics 2014; 15: 732.
Wang C, Evans JM, Bhagwate AV, Prodduturi N, Sarangi V, Middha M et al. PatternCNV: a versatile tool for detecting copy number changes from exome sequencing data. Bioinformatics 2014; 30: 2678–2680.
Huang da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Liu T, Papagiannakopoulos T, Puskar K, Qi S, Santiago F, Clay W et al. Detection of a microRNA signal in an in vivo expression set of mRNAs. PLoS ONE 2007; 2: e804.
Gautier L, Cope L, Bolstad BM, Irizarry RA . affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307–315.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 2014; 26: 288–300.
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M . Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014; 42: D199–D205.
Jiao X, Sherman BT, Huang da W, Stephens R, Baseler MW, Lane HC et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012; 28: 1805–1806.
Watanabe M, Kawaguchi T, Isa S, Ando M, Tamiya A, Kubo A et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res 2015; 21: 3552–3560.
McDonald OG, Wu H, Timp W, Doi A, Feinberg AP . Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struc Mol Biol 2011; 18: 867–874.
De Craene B, Berx G . Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110.
Anastassiou D, Rumjantseva V, Cheng W, Huang J, Canoll PD, Yamashiro DJ et al. Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo. BMC Cancer 2011; 11: 529.
Jechlinger M, Grunert S, Tamir IH, Janda E, Ludemann S, Waerner T et al. Expression profiling of epithelial plasticity in tumor progression. Oncogene 2003; 22: 7155–7169.
Thiery JP, Acloque H, Huang RYJ, Nieto MA . Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110.
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360: 765–773.
Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res 2004; 64: 6892–6899.
Fujisawa H, Reis RM, Nakamura M, Colella S, Yonekawa Y, Kleihues P et al. Loss of heterozygosity on chromosome 10 is more extensive in primary (de novo) than in secondary glioblastomas. Lab Invest 2000; 80: 65–72.
Tohma Y, Gratas C, Biernat W, Peraud A, Fukuda M, Yonekawa Y et al. PTEN (MMAC1) mutations are frequent in primary glioblastomas (de novo) but not in secondary glioblastomas. J Neuropathol Exp Neurol 1998; 57: 684–689.
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006; 9: 157–173.
Tam WL, Weinberg RA . The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013; 19: 1438–1449.
Nagaishi M, Paulus W, Brokinkel B, Vital A, Tanaka Y, Nakazato Y et al. Transcriptional factors for epithelial-mesenchymal transition are associated with mesenchymal differentiation in gliosarcoma. Brain Pathol 2012; 22: 670–676.
Lamouille S, Xu J, Derynck R . Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196.
Davis FM, Azimi I, Faville RA, Peters AA, Jalink K, Putney JW Jr et al. Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent. Oncogene 2014; 33: 2307–2316.
Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011; 13: 317–323.
Ananda G, Mockus S, Lundquist M, Spotlow V, Simons A, Mitchell T et al. Development and validation of the JAX Cancer Treatment Profile (TM) for detection of clinically actionable mutations in solid tumors. Exp Mol Pathol 2015; 98: 106–112.
Acknowledgements
We thank Y Dorsett for comments on this manuscript. This work was supported in part by grants to CL, an Ewha Womans University Research Grant; the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI13C2148); and the International Research & Development Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (grant no. 2015K1A4A3047851). Grants to SHP, including the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI11C21100200); the Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries, Republic of Korea (grant no. 311011-05-3-SB020); the Technology Innovation Program (grant no. 10050154, Business Model Development for Personalized Medicine Based on Integrated Genome and Clinical Information) funded by the Ministry of Trade, Industry & Energy (MI, Korea); and the Bio & Medical Technology Development Program of the NRF funded by the Korean government, MSIP (grant no. 2015M3C7A1028926). A grant to D-HN included the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI14C3418).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Cho, SY., Park, C., Na, D. et al. High prevalence of TP53 mutations is associated with poor survival and an EMT signature in gliosarcoma patients. Exp Mol Med 49, e317 (2017). https://doi.org/10.1038/emm.2017.9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2017.9
This article is cited by
-
Prevalence, causes and impact of TP53-loss phenocopying events in human tumors
BMC Biology (2023)
-
Primary gliosarcoma with widespread extracranial metastases—spatiotemporal morphological variation
Chinese Neurosurgical Journal (2022)
-
Mutated RAS-associating proteins and ERK activation in relapse/refractory diffuse large B cell lymphoma
Scientific Reports (2022)
-
Gliosarcoma vs. glioblastoma: a retrospective case series using molecular profiling
BMC Neurology (2021)
-
Hypermutated phenotype in gliosarcoma of the spinal cord
npj Precision Oncology (2021)