Abstract
A karyotype of Trifolium repens constructed from mitotic cells revealed 13 pairs of metacentric and 3 pairs of submetacentric chromosomes including a pair of satellites located at the end of the short arm of chromosome 16. C-bands were identified around the centromeric regions of 8 pairs of chromosomes. A 350 bp tandemly repeated DNA sequence from T. repens labelled with digoxygenin hybridized to the proximal centromeric regions of 12 chromosome pairs. Some correlation between the distribution of the repeat sequence and the distribution of C-banding was demonstrated.
Similar content being viewed by others
Introduction
Trifolium repens L, white clover, is an economically important plant species in temperate pastures. As briefly reported by1, it has 16 pairs of chromosomes (2n = 32). As yet, no detailed cytological examination of this species, such as C-banding, has been reported. In the last decade, the technique of C-banding has been used to examine highly repeated sequences in plant chromosomes and has provided a useful tool for the analysis of cytogenetic structure in crop plants 2, 3, 4, 5, 6, 7.
In plants, the chromosomal localization of highly repeated DNA sequences by in situ hybridization using non-radioactive labelling and detection techniques has been reported for cereals8, 9, 10, 11, 12, Allium13, tomato14, 15, 16, rice17, soybean18, Picea19, Festuca20, and Arabidopsis21.
For this study, a cytological examination of T. repens chromosomes from root tips was undertaken to obtain a basic karyogram using conventional Giemsa staining as well as more detailed C-banding information. In addition, a tandemly repeated, 350 bp, T. repens DNA sequence labelled with digoxygenin was used for an in situ hybridization analysis of T. repens chromosomes.
Materials and Methods
Plant material and chromosome preparation
Seeds of T. repens cv. Huia were obtained from the Margot Forde Germplasm Centre (New Zealand Pastoral Agriculture Research Institute, Grasslands Research Centre, Palmerston North, New Zealand). Seeds were germinated at room temperature for 2-3 d in a moist petri dish. Root tips of 2-3 d old seedlings were treated with either a saturated solution of paradichlorobenzene (BDH) for 2-3 h or 0.05% colchicine (BDH) for 1-2 h and then fixed in a freshly prepared solution of 1:3 acetic acid/ethanol overnight at 4 °C. Chromosome preparations were carried out according to the procedure of Zhu and Gardiner22. Briefly, the enzyme treatment was used to weaken cell walls of root tips. Partially digested root tips were rinsed in distilled water and then placed in a drop of 45% acetic acid on a glass slide. The meristematic region of the root tip was excised teased out. Cells and chromosomes were gently squashed under a glass cover slip on a warm plate and stored at room temperature for use.
C-banding
C-banding was carried out according to the procedure of Zhu and Gardiner23. Slides were examined under a microscope (Zeiss) with a 100×oil objective. Mitotic spreads displaying sharp C-bands were photographed on Kodak Technique Pan black and white 2415 film.
DNA probe and in situ hybridization
Clone pTr350.1 (EMBL Accession No. X58701) containing a 350 bp tandemly repeated T. repens DNA sequence element was subcloned from a lambda genomic clone with an approx, llkb insert consisting entirely of monomeric 350 bp and dimeric 700 bp repeating elements (data not shown). The insert from clone pTr350.1 was labelled with digoxygenin-ll-dUTP using random primer labelling according to the manufacturer's specifications (Boehringer Mannheim).
In situ hybridization was carried out according to the procedure of Zhu and Gardiner22. Hybridization sites were detected by an alkaline phosphatase-conjugated antibody according to the manufacturer's instructions for the labelling and detection kit (Boehringer Mannheim). Visualization of the bound antibody was accomplished by an enzymatic colour reaction, the colourometric substrates nitroblue tetrazolium salt (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) producing a dark purple/brown precipitate at the sites of hybridization on the chromosome. Controls consisted of a chromosome spread pretreated with 50 μg/ml DNase (Sigma) at 37 °C for 30 min and a chromosome spread carried through the hybridization procedure but without hybridization of a probe.
Measurements were made from photographs of 11 mitotic spreads. Relative chromosome lengths and arm ratios were calculated according to Macgregor and Varley24. Karyotypes of chromosomes were made from enlarged photomicrographs with the chromosomes arranged in order of decreasing length. The chromosomes were then compared on the basis of centromere position, arm ratio, and the sizes of the satellites.
Results and Discussion
Cytological analysis
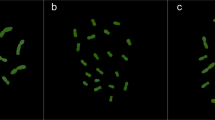
T. repens has 16 pairs of chromosomes with a pair of satellites located at the end of the short arm of chromosome 16 (Fig 1), confirming the earlier results reported by Chen and Gibson1. A somatic chromosome karyotype was constructed from 11 mitotic cells by arranging the chromosome pairs on the basis of decreasing size and centromere position, with classification on the basis of the arm ratio using the criteria of Levan et al.25 (Tab 1). Karyotype analysis showed that 13 pairs of chromosomes are metacentric (chromosomes 1, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, and 15) and 3 pairs are submetacentric (chromosomes 2, 13, and 16). In contrast, Chen and Gibson1, in a less detailed analysis, reported that 4 pairs of the T. repens chromosomes were metacentric, 11 pairs were submetacentric, and 1 pair was telocentric.
C-banding analysis
C-bands were observed at the proximal centromeric regions of chromosome pairs 2, 3, 5, 7, 9, ll, 13, and 15 while the other 8 pairs of chromosomes lacked.C-bands (Fig 2). Neither terminal nor interstitial C-bands were observed. This C-banding pattern was also observed by Dr H. Ansari (personal communication). Giemsa C-banding is associated with highly repeated DNA sequences in constitutive heterochromatin24. The C-banding technique has been used to visualize the cytogenetic condition and the chromosomal composition of plants such as cereals2, 4, 5, 6, 11, 26, 27 and Vicia species28.
In situ hybridization
The digoxygenin-labelled 350 bp T. repens repeat DNA hybridized to 12 pairs of chromosomes (chromosome pairs 1, 2, 3, 5, 6, 7, 9, 11, 12, 13, 15, and 16) at proximal centromeric regions (Fig 3) with a 90% frequency based on 11 karyotypes (data not shown). In all cases the hybridization signals were strong and could even be observed at late prophase (Fig 4). A low frequency of apparent hybridization (<27%) at the proximal centromeric regions of chromosomes 4, 8, 10, and 14 was observed. The hybridization signals on the small chromosomes of T. repens obtained using the digoxygenin-labelled probe were considerably more precise and distinct than those obtained using radioactively-labelled probes (data not shown).
As for Fig 3 but at late prophase.
Three classes of tandemly repeated DNA sequences in human chromosomes have been identified, i.e. classical satellite, α-satellite, and β-satellite DNA29. All of these sequences are located in the variable heterochromatic regions on particular chromosomes30. The nucleotide sequence and the organization of monomeric units varies from one chromosome to another in the form of chromosome-specific, higher order, tandemly repeated arrays (alphoid subfamilies)29.
Alphoid-like satellite DNA sequences have been characterised for a number of plant species including radish31, cereals9, and members of the Liliaceae32. In all cases these sequences were located in the variable heterochromatic regions of specific chromosomes. For T. repens, the 350 bp repeat DNA was found to be localized in heterochromatin around the centromeric regions on specific chromosomes and appeared to be one component of the centromeric DNA of this species. Chromosomes 4, 8, 10, and 14, which did not exhibit, or exhibit only a low frequency of hybridization with this probe, may have different and/or variable satellite DNA sequences involved in these centromeric regions as is the case in humans and cereals as discussed above. Alternatively, since T. repens is tetraploid, this may reflect differences within the genomes of the respective parental species.
For T. repens, the C-banding technique has revealed the presence of heterochromatin around the centromeric regions of some chromosomes. There is some similarity between the patterns of labelling revealed by in situ hybridization with the 350 bp repeat DNA and the C-banding pattern. Chromosome pairs 1, 6, and 13 (Fig 3) show obvious in situ hybridization signals where there are no apparent C-bands, probably reflecting the greater loss of some nucleic acid during C-banding procedure and reflecting the more sensitivity of the in situ hybridization technique. However, chromosome pairs 2, 7, and 9 all show clear hybridization of the 350 bp repeat DNA to both homologues of each pair of chromosomes yet only one of each of the homologues exhibits C-banding. This phenomenon needs to be further investigated.
Additional future work could involve an investigation into the distribution of the 350 bp repeat sequences of T. repens in closely related clover species. The use of fluorescent staining33 and fluorescent in situ hybridization techniques using the 350 bp repeat sequence probe, along with other probes, could also be considered in order to analyse the chromosomal structure of T. repens in greater detail.
Accession codes
References
Chen CC, Gibson PB . Karyotypes of fifteen Trifolium species in section Amoria. Crop Science 1971; 1:441–5.
Seal AG . C-banded wheat chromosomes in wheat and triticale. Theor Appl Genet 1982; 63:39–47.
Lapitan NLV, Sears RG, Rayburn AL, Gill BS . Wheat-rye translocations. J Hered 1986; 77:415–9.
Linde-Laursen IB, Bothmer R . Giemsa C-banding in two polyploid, South American Hordeum species, H. tetraploidum and H. lechleri, and their aneuploid hybrids with H. vulgare. Hereditas 1986; 105:171–7.
Hainer A, Hesemann CU . Cytogenetic investigations in rye, wheat and triticale. 3. C-banding of tetra- and hexaploid wheat by Giemsa- and/or Leishman staining. Theor Appl Genet 1988; 75:305–8.
Gill BS, Friebe B, Endo TR . Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 1991; 34:830–9.
Cai X . Chromosome translocations in the common wheat variety 'Amigo'. Hereditas 1994; 121:199–202.
Rayburn AL, Gill BS . Use of biotin-labeled probes to map specific DNA sequences on wheat chromosomes. J Hered 1985; 76:78–81.
Rayburn AL, Gill BS . Use of repeated DNA sequences as cytological markers. Amer J Bot 1987; 74:574–80.
Mukai Y, Endo TR, Gill BS . Physical mapping of the 18S.26S rRNA multigene family in common wheat: Identification of a new locus. Chromosoma 1991; 100:71–8.
Hohmann U, Lagudah ES . C-banding polymorphism and linkage of nonhomoeologous RFLP loci in the D genome progenitor of wheat. Genome 1993; 36:235–43.
Nkongolo KK, Lapitan NLV, Quick JS, Muhlmann MD . An optimized fluorescence in situ hybridization procedure for detecting rye chromosomes in wheat. Genome 1993; 36:701–5.
Ricroch A, Peffley EB, Baker RJ . Chromosomal location of rDNA in Allium: in situ hybridiza- tion using biotin- and fluorescein-labelled probe. Theor Appl Genet 1992; 83:413–8.
Lapitan NLV, Ganal MW, Tanksley SD . Somatic chromosome karyotype of tomato based on in situ hybridization of the TGRI satellite repeat. Genome 1989; 32:992–8.
Lapitan NLV, Ganal MW, Tanksley SD . Organization of the 5S ribosomal RNA genes in the genome of tomato. Genome 1991; 34:509–14.
Ganal MW, Lapitan NLV, Tanksley SD . A molecular and cytogenetic survey of major repeated DNA sequences in tomato (Lycopersicon esculentum). Mol Gen Genet 1988; 213:262–8.
Song YC, Gustafson JP . Physical mapping of the 5S rDNA gene complex in rice (Oryza sativa). Genome 1993; 36:658–61.
Griffor MC, Vodkin LO, Singh RJ, Hymowitz T . Fluorescent in situ hybridization to soybean metaphase chromosomes. Plant Mol Biol 1991; 17:101–9.
Brown GR, Amarasinghe V, Kiss G, Carlson JE . Preliminary karyotype and chromosomal loca- lization of ribosomal DNA sites in white spruce using fluorescence in situ hybridization. Genome 1993; 36:310–6.
Bailey JP, Bennett ST, Bennett MD, Stace CA . Genomic in situ hybridization identifies parental chromosomes in the wild grass hybrid × Festulpia Hubbardii Heredity, 1993; 71:413–20.
Maluszynska J, Heslop-Harrison JS . Localization of tandemly repeated DNA sequences in Ara- bidopsis thaliana. Plant J 1991; 1:159–66.
Zhu JM, Gardiner SE . Chromosomal localization of ribosomal DNA sequeces in an apple root- stock using a digoxygenin detection system. Cell Res 1995; 5:1–7
Zhu JM, Gardiner SE, Lay-Yee M . Physical mapping of three fruit ripening genes: Endopoly- galacturonase ACC oxidase and ACC synthase from apple (Malus×domestica) in an apple rootstock A106 (Malus sieboldii). Cell Res 1995; 5:243–54.
Macgregor HC, Varley JM . Working with animal chromosomes. John Wiley and Sons Ltd, Chichester: 1983; 250pp.
Levan A, Fredga K, Sandberg AA . Nomenclature for centromeric position on chromosomes. Hereditas 1964; 52:201–20.
Rayburn AL, Gill BS . Molecular identification of the D-genome chromosomes of wheat. J Hered 1986; 77:253–5.
Stöβer T, Günther T, Hesemann CU . Banding of rye (Secale cereale) chromosomes using the restriction enzymes Alu I, Dra I, Hpa II, and Msp I. Genome 1993; 36:998–1002.
Cremonini R, Funari S, Galasso I, Pignone D . Cytology of Vicia species II. Banding patterns and chromatin organization in Vicia atropurpurea Desf. Heredity 1993; 70:628–33.
Lin CC, Sasi R, Lee C, Fan YS, Court D . Isolation and identification of a novel tandemly repeated DNA sequence in the centromeric region of human chromosome 8. Chromosoma 1993; 102:333–9.
Heng HHQ, Tsui LC . Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma 1993; 102:325–32.
Grellet F, Delcasso D, Panabieres F, Delseny M . Organization and evolution of a higher plant alphoid-like satellite DNA sequence. J Mol Biol 1986; 187:495–507.
Deumling B, Greilhuber J . Characterization of heterochromatin in different species of Scilla siberica group (Liliaceae) by in situ hybridization of satellite DNAs and fluorochrome banding. Chromosoma 1982; 84:535–55.
Guerra M . Cytogenetics of Rutaceae. V. High Chromosomal variability in Citrus species revealed by CMA/DAPI staining. Heredity 1993; 71:234–41.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhu, J., Ellison, N. & Rowland, R. Chromosomal localization of a tandemly repeated DNA sequence in Trifolium repens L.. Cell Res 6, 39–46 (1996). https://doi.org/10.1038/cr.1996.5
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1996.5