Abstract
Necrosis leads to the release of so-called damage-associated molecular patterns (DAMPs), which may provoke inflammatory responses. However, the release of organelles from dying cells, and the consequences thereof have not been documented before. We demonstrate here that mitochondria are released from cells undergoing tumor necrosis factor-α (TNF-α)-induced, receptor-interacting protein (RIP)1-dependent necroptosis, a form of programmed necrosis. The released, purified mitochondria were determined to be intact as they did not emit appreciable amounts of mitochondrial DNA (mtDNA). Pharmacological inhibition of dynamin-related protein 1 (Drp1) prevented mitochondrial fission in TNF-α-triggered cells, but this did not block necroptosis nor the concomitant release of mitochondria. Importantly, primary human macrophages and dendritic cells engulfed mitochondria from necroptotic cells leading to modulation of macrophage secretion of cytokines and induction of dendritic cell maturation. Our results show that intact mitochondria are released from necroptotic cells and suggest that these organelles act as bona fide danger signals.
Similar content being viewed by others
Main
Dying cells need to be cleared under physiological conditions, as they may otherwise release noxious contents causing inflammation and tissue damage. The inefficient disposal of dying cells or cell debris may also elicit autoimmune responses.1 The mechanisms of clearance of apoptotic cells by phagocytes appear to be largely conserved through evolution. It has thus been shown that apoptotic cells in mammals and nematodes express ‘eat-me’ signals such as phosphatidylserine (PS) on the cell surface to trigger recognition by phagocytes.2 This leads to the swift and ‘silent’, that is, non-inflammatory removal of apoptotic cells. In recent years, a form of regulated necrosis, so-called necroptosis, has been described.3, 4 Necroptosis is typically initiated via death receptors, such as Fas or TNF receptor, leading to the activation of receptor-interacting protein kinase 1 or 3 (RIP1/RIP3). Although the signaling pathways underlying the execution of necroptosis are coming to light,5 the clearance of necroptotic cells, and the subsequent outcomes of necroptotic cell death, is not well understood. Indeed, necroptosis may result in the immunologically silent maintenance of immune homeostasis or, alternatively, may provoke strong inflammatory responses, which may be coupled to the emission of ‘danger’ signals from necroptotic cells (for an excellent review, see Kaczmarek et al.6).
Microbial pathogen-associated molecular patterns (PAMPs) activate the immune system. Similarly, cellular injury may cause the release of damage-associated molecular patterns (DAMPs) to activate the innate immune system. Zhang et al.7 reported that severe trauma releases mitochondrial DAMPs (mtDAMPs) such as mitochondrial DNA (mtDNA) and formyl peptides into the circulation, and that these factors signal through innate immune pathways identical to those activated in sepsis. Furthermore, intravenous injection of mtDAMPs resulted in marked inflammatory lung injury in rats.7 Collins et al.8 detected extracellular mtDNA in the synovial fluids of rheumatoid arthritis patients. Iyer et al.9 provided evidence that necrotic (pressure-disrupted) cells are sensed by the NLRP3 inflammasome resulting in release of pro-inflammatory interleukin (IL)-1β. This activation was triggered in part through release of ATP produced by mitochondria released from damaged cells. As pointed out recently by Masters and Walsh,10 the latter observations raise interesting questions concerning the nature of sterile inflammation, and supports an evolutionarily conserved link between pathogenic bacteria and (endosymbiont) mitochondria. Nonetheless, it remains unclear whether intact mitochondria are released from dying cells and whether these organelles act as danger signals. In the present study, using established in vitro models of necroptosis, we investigated whether mitochondria are released during cell death and whether they are recognized by immune cells.
Results
TNF-α induces necroptosis in FADD-deficient Jurkat cells and L929 cells
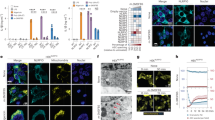
To study necroptosis, we used Fas-associated protein with death domain (FADD)-deficient Jurkat (human T-lymphoblastic leukemia) and L929 (murine fibroblast) cells treated with tumor necrosis factor-α (TNF-α), which is known to induce necroptosis via RIP1/RIP3 activation.11 The small-molecule inhibitor, necrostatin-1 (Nec-1) specifically blocks this cell death.4 Consistently, although Jurkat wild-type cells did not show PS exposure upon TNF-α stimulation (Figure 1a), FADD-deficient Jurkat cells and L929 cells displayed PS exposure after 24 h, and this was inhibited by Nec-1, but not by zVAD-fmk, a pan-caspase inhibitor known to block apoptosis (Figures 1a–c). The morphology of necroptotic, FADD-deficient Jurkat cells was observed using transmission electron microscopy (TEM) (Figure 1d). Compared with non-treated cells possessing normal mitochondrial morphology, TNF-α-treated cells showed a disrupted plasma membrane and disappearance of intracellular components, typical of necrotic cells. Interestingly, we could observe small vesicles adjacent to the plasma membrane. These vesicles were around 500 nm in diameter, which is similar to the size of mitochondria in control cells. We also performed flow cytometry-based sorting of MitoTracker-positive events, and subjected the pellet to TEM imaging. As shown in Figure 2a, we could observe intact mitochondria albeit with a swollen appearance, with characteristic cristae structures. The latter finding is in line with previous work by Degterev et al.3 who noted swelling of mitochondria (within dying cells) during necroptosis. Furthermore, the presence of mitochondria in the pellet derived from the supernatants of necroptotic cells was confirmed by western blot using anti-cytochrome c oxidase IV (COX-IV) antibody (Figure 2b). Mitochondria purified from TNF-α-treated FADD-deficient Jurkat cells (hereafter referred to as mito-pure) were included as a control. These mitochondria were also shown to be MitoTracker-positive by flow cytometry (Figure 2c) (and see below).
TNF-α induces RIP1/RIP3-dependent necroptosis. (a) Wild-type or FADD-deficient Jurkat cells were treated with either 40 μM Nec-1 or 10 ng/ml TNF-α, or both, for 24 h. Cells were stained with annexin V-FITC and analyzed by flow cytometry. FADD-deficient Jurkat cells (b) and L929 cells (c) were treated with 10 ng/ml TNF-α in the presence of 40 μM (for Jurkat cells) or 10 μM (for L929 cells) Nec-1 (RIP1/RIP3 inhibitor) or 20 μM zVAD-fmk (pan-caspase inhibitor) for 24 h. Cells were stained with annexin V-FITC as above. (d) TEM pictures of FADD-deficient Jurkat cells (control, upper panels; or TNF-α-treated for 24 h, lower panels). Note the presence of extracellular, mitochondria-like vesicles adjacent to the plasma membrane following necroptosis
TNF-α induces extracellular release of mitochondria. (a) The pellet collected from TNF-α-treated FADD-deficient Jurkat cells was stained with MitoTracker Green and then sorted by flow cytometry based on MitoTracker Green-positive events. The collected events/particles were visualized by TEM. Two representative images are shown. Note the presence of cristae-like structures. Scale bar: 200 nm. (b) The crude pellets from the cell culture supernatant of TNF-α-treated FADD-deficient Jurkat cells incubated in presence or absence of 40 μM Nec-1 and purified mitochondria (mito-pure) from TNF-α-treated FADD-deficient Jurkat cells were suspended in the same volume of buffer and monitored by western blot with anti-COX-IV antibody. (c) Purified mitochondria were stained with MitoTracker Green and analyzed by flow cytometry. The depicted histograms correspond to non-stained and MitoTracker-stained mito-pure
TNF-α induces mitochondrial fission and extracellular release of mitochondria
Next, plasma membrane disruption of cells undergoing necroptosis was monitored using the vital dye, trypan blue. Trypan blue-positive cells increased in a time-dependent manner reaching a plateau at around 12 h after TNF-α treatment, and this was blocked by Nec-1 (Figure 3a). To assess the mitochondrial content in cells, we performed western blotting for COX-IV and noted a decrease of mitochondrial protein at 9 h after TNF-α treatment. This was prevented by Nec-1 confirming that the change was related to necroptosis (Figure 3b). To further support this result, we monitored the mitochondrial content by time-lapse confocal imaging upon TNF-α stimulation using the specific dye, MitoTracker Green. After 6 h, mitochondrial staining was reduced and a dot-like pattern suggestive of mitochondrial fission was noted in the FADD-deficient Jurkat cells (Figure 3c). We observed a similar change in mitochondrial morphology in L929 cells after 6 h of treatment with TNF-α (Figure 3d). Notably, propidium iodide (PI) staining of the cell nuclei of FADD-deficient Jurkat cells was evident at 7 h and onward. At this time, the MitoTracker staining was no longer detectable. It thus appears that the loss of mitochondrial staining during TNF-α-triggered necroptosis occurs before the disruption of the plasma membrane. This implies that the reduction of mitochondrial content—indicative of release of mitochondria from necroptotic cells—may transpire in a regulated manner. To further support that loss of mitochondrial staining is due to the release of mitochondria, we collected cell pellets from the cell culture supernatant of FADD-deficient Jurkat cells (Figure 3e) and L929 cells (Figure 4c) treated or not treated with TNF-α, and stained the pellets with MitoTracker Green. The pellet from TNF-α-treated FADD-deficient Jurkat cells displayed an increased number of MitoTracker-positive events and this was reduced in Nec-1-treated cells (Figure 3e). Similar results were obtained for L929 cells (Figure 4c). Taken together, we provide evidence for the release of mitochondria during necroptosis. Furthermore, the released mitochondria appeared to be morphologically intact (Figure 2a), and experiments described below suggest that released mitochondria do not ‘leak’ mtDNA.
TNF-α induces early release of mitochondria during necroptosis. (a) FADD-deficient Jurkat cells were treated with 10 ng/ml of TNF-α with/without 40 μM Nec-1, and stained with trypan blue. Data shown are from three independent experiments. (b) FADD-deficient Jurkat cells were treated with 10 ng/ml of TNF-α with/without 40 μM Nec-1 for the indicated time and cell lysates were blotted with anti-COX-IV antibody. β-actin antibody is used as loading control. (c) Time-lapse imaging of TNF-α-treated FADD-deficient Jurkat cells. Cells were treated with 10 ng/ml TNF-α in the presence of MitoTracker Green and propidium iodide (red). (d) Fluorescence microscopic images of L929 cells treated or not with 5 ng/ml of TNF-α for 6 h. Cells were stained with MitoTracker Deep Red and DAPI to visualize the cell nucleus. Scale bars: 10 μm. (e) The pellets were collected from the supernatant of the FADD-deficient Jurkat cells cultured medium after 24 h incubation with 10 ng/ml of TNF-α with/without 40 μM Nec-1, and the crude pellet was stained with MitoTracker Green. The fluorescence-labeled pellets were analyzed by flow cytometry. The graph shows the fold increase of MitoTracker-positive events when compared with the control. Data shown are mean values±S.E.M. of three independent experiments. ***P<0.001 by the Student’s t-test
Mitochondrial fission and release during TNF-α-mediated necroptosis. (a) L929 cells were treated with the indicated doses of Mdivi-1 or 10 μM Nec-1 for 6 h and then treated with or without 5 ng/ml of TNF-α for 12 h. The cells were stained with propidium iodide and analyzed by flow cytometry. Data shown are mean values±S.E.M. of three independent experiments. (b) Fluorescence microscopic image of L929 cells pre-treated with 20 μM Mdivi-1 for 6 h and then treated or not with TNF-α for 6 h. Then, cells were stained with MitoTracker Deep Red and DAPI. Note the shortening of mitochondria (see inset for higher magnification). Scale bars: 10 μm. (c) The pellets were collected from the cell culture supernatant of L929 cells after 12 h incubation with TNF-α with/without pre-treatment with 20 μM Mdivi-1 for 6 h or in the presence of 10 μM Nec-1, and then stained with MitoTracker Green. The fluorescence-labeled pellets were analyzed by flow cytometry. The graphs depict the fold increase of MitoTracker-positive events compared with the control sample. Data shown are the mean values±S.E.M. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 by the Student’s t-test
Inhibition of Drp1 promotes necroptosis and the release of mitochondria
Using a combination of TNF-α, z-VAD-fmk and a Smac mimetic to trigger necroptosis, Wang et al.12 reported that mitochondrial fragmentation is an early and obligatory step for necrosis execution. In light of the morphological appearance of mitochondria in cells undergoing necroptosis in our study (Figures 3c and d), we hypothesized that mitochondrial fission may be related to necroptotic cell death and the release of these organelles. To address whether the mitochondrial fission factor, Drp1, is required for necroptosis in our model, we used the compound Mdivi-1 that acts as an inhibitor of Drp1.13 Interestingly, Mdivi-1 treatment did not inhibit TNF-α-induced cell death of L929 cells (Figure 4a), although mitochondrial fission was blocked, as evidenced by the presence of elongated mitochondria (Figure 4b). In fact, co-treatment of cells with Mdivi-1 and TNF-α resulted in increased cell death, which was prevented by Nec-1 (Figure 4a), suggesting that inhibition of mitochondrial fission enhances necroptotic cell death. Moreover, as we examined the amount of released mitochondria from cells co-treated with Mdivi-1 and TNF-α, we observed an increased release of mitochondria when compared with cells treated only with TNF-α (Figure 4c), suggesting that release of mitochondria in cells undergoing TNF-α-induced necroptosis is potentiated by inhibition of Drp1.
Human innate immune cells recognize released mitochondria as DAMPs
We hypothesized that the release of mitochondria—cellular organelles that are thought to have originated as symbiotic bacteria14—could serve to activate immune-competent cells. We therefore purified the mitochondria released from necroptotic, FADD-deficient Jurkat cells and examined whether these released mitochondria are engulfed by primary human monocyte-derived macrophages (HMDMs), key scavengers of dying cells and cell debris.15 As shown in Figure 5a, mito-pure were taken up by HMDMs, and this was inhibited by cytochalasin D, indicating that the uptake occurred through an active, cytoskeleton-dependent process. The cellular internalization of mito-pure was confirmed by confocal microscopy. Mito-pure pre-stained with MitoTracker Green were thus observed as small dots inside the cells (Figure 5b). Moreover, primary human monocyte-derived dendritic cells (MDDCs), professional antigen-presenting cells of the immune system,15 were also found to engulf mito-pure (Figure 5b). To examine whether human macrophages respond to mito-pure, we performed cytokine measurements on supernatants of HMDMs exposed to mito-pure for 6 h. Nine cytokines (TNF-α, IL-1β, IL-6, IL-12p40, IL-18, IFN-α2, IL-4, IL-8, and IL-10) were evaluated using a multiplex array, and we were able to detect four of these cytokines in cell culture supernatants (TNF-α, IL-6, IL-8, and IL-10), but the level of expression of the remaining cytokines was below the detection limit. Lipopolysaccharide (LPS), a component of the outer membrane of Gram-negative bacteria, was included as a positive control. We thus detected a modest albeit significant induction of the pro-inflammatory cytokines, TNF-α and IL-6 and—similarly—a dose-dependent induction of the immunomodulatory cytokine, IL-10, in macrophages in response to mito-pure (Figure 5c). Mito-pure also triggered a pronounced (i.e., comparable to LPS) induction of IL-8 (recently re-named CXCL8), a pro-inflammatory mediator that induces chemotaxis in target cells, mainly neutrophils. In addition, we found that incubation of primary human MDDCs with mito-pure resulted in maturation of these cells, as evidenced by the induction of the cell surface markers, CD80, CD83, and CD86 (Figure 5d). LPS was used as a positive control.
Mitochondria released by necroptotic cells act as danger signals. (a) HMDMs were incubated with MitoTracker-labeled, purified mitochondria (mito-pure) for 1 h in HBSS. After incubation, cells were analyzed by flow cytometry to determine the percentage of phagocytosis-positive macrophages. Cells were pre-treated with cytochalasin D for 30 min before adding mito-pure. Data represent mean values±S.E.M. of three independent experiments. (b) HMDMs or MDDCs incubated for 1 h with 5 μg mito-pure pre-stained with MitoTracker Green were stained with Phalloidin and DAPI, and then observed by confocal microscopy. Scale bars: 20 μm. (c) HMDMs were co-incubated with mito-pure for 1 h in HBSS, and then for 5 h in fresh medium. Cell culture supernatants were collected and the level of cytokines was determined as detailed in Materials and Methods. Treatment with 100 ng/ml LPS for 6 h was included as a positive control. Data are shown as mean values±S.E.M. of three independent experiments. (d) MDDCs were co-incubated with mito-pure for 24 h, then cells were stained with the indicated antibodies (CD80, CD83, or CD86) conjugated with FITC. Relative expression of surface markers was determined by flow cytometry. Data shown are mean values±S.E.M. of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 by the Student’s t-test
Finally, to verify that the mitochondria released from necroptotic cells were intact, we performed real-time PCR analysis of the buffer in which mito-pure had been incubated (Figure 6a). Trace amounts (about 1.5 ng/ml) of mtDNA were detected in the buffer incubated for 1 h with mito-pure, and a similar trace amount was present at 0 h, suggesting that this is owing to the procedure used for purification of mitochondria. More importantly, this amount of mtDNA is a 1000-fold lower than the amount that was shown to induce cytokine release in neutrophils (1–10 μg/ml) in previous studies of mtDAMPs.7 Nonetheless, to further assess whether mtDNA could affect cytokine production in HMDMs, we extracted mtDNA from mitochondria purified from FADD-deficient Jurkat cells, and exposed HMDMs to mtDNA alone (1–10 000 ng/ml) or in combination with LPS (100 ng/ml) followed by the detection of cytokines in the cell culture supernatants. mtDNA had no effect on macrophage production of TNF-α (Supplementary Figure 1) or IL-8 (data not shown). Similar experiments were performed using primary human neutrophils, but we could not detect any secretion of cytokines by cells exposed to mtDNA alone, nor did we observe any potentiation of TNF-α or IL-8 secretion in neutrophils exposed to mtDNA in combination with formyl-Met-Leu-Phe (fMLP) versus fMLP alone (data not shown).
Necroptotic cell-derived mitochondria are intact. (a) Mitochondria purified from necroptotic cells (mito-pure) were incubated in HBSS for 0 or 1 h at 37 °C and the buffer was then collected and pellets were removed by centrifugation. The samples were analyzed by RT-PCR for the mitochondrial gene, Co1. Purified mtDNA from mito-pure was used as input (4 μg/ml) and a standard curve was generated to quantify the amount of mtDNA. Note that only trace amounts of mtDNA (in the range of 1.5 ng/ml) could be detected in the supernatants of the mito-pure samples. (b) Schematic representation of (upper panel) release of mtDAMPs during trauma, resulting in neutrophil activation, with migration and degranulation,7 and (lower panel) release of intact mitochondria during TNF-α-triggered necroptosis, leading to secretion of pro-inflammatory cytokines by macrophages and dendritic cell maturation. According to these two scenarios, mtDAMPs (such as mtDNA or formyl peptides) as well as intact mitochondria can act as ‘danger’ signals
Discussion
We have shown herein that mitochondria are released from cells undergoing TNF-α-induced, RIP1-dependent necroptosis, a form of programmed necrosis. Importantly, we found that primary human macrophages and DCs are able to engulf mitochondria released from necroptotic cells, and that these organelles act as danger signals, as evidenced by their effect on cytokine production (by macrophages) and cell maturation (of antigen-presenting DCs). Intact nuclei have been observed in the extracellular milieu during necroptosis;16 however, whether this is a result of the disintegration of dying cells or due to active expulsion of cell nuclei was not clarified. The present study provides evidence for the active release of intact organelles, and our cytokine profiling data suggest that these are ‘sensed’ as pathogens, which is in accordance with the notion that mitochondria have originated as symbiotic bacteria.14 Zhang et al.7 reported that severe trauma releases mtDNA and formyl peptides into the circulation with functionally important immune consequences. Hence, these so-called mtDAMPs were found to activate neutrophils, leading to neutrophil migration and degranulation in vitro and in vivo, and circulating mtDAMPs were suggested to elicit neutrophil-mediated organ injury.7 In a subsequent study, mtDAMPs were implicated both in neutrophil-dependent and -independent pathways of endothelial permeability during systemic inflammation,17 suggesting that mtDAMPs may be important therapeutic targets. Cellular DAMPs such as high-mobility group box-1 are thought to be released by necrotic cells, but not by apoptotic cells.18 However, Wickman et al.19 have recently shown that the membranes of apoptotic bodies emanating from apoptotic cells do not form impermeable barriers thus allowing for the egress of macromolecules before cells undergo ‘secondary necrosis’. Using quantitative proteomics approaches, validated DAMPs were shown to be among the proteins that are released from apoptotic bodies upon TNF-α plus cycloheximide treatment.19 In the present study, TNF-α-induced necroptosis is shown to be accompanied by the release of mitochondria before the disruption of the plasma membrane, indicating that this is an active process; moreover, the released mitochondria from necroptotic cells were shown to be intact, as no mtDNA was emitted from the purified, released mitochondria. Whether or not these ‘necroptotic’ mitochondria remain functionally active after their extracellular release and/or following internalization by phagocytic cells was not assessed in the present study, but this could be an interesting point to consider in the future. Notably, transfer of functionally active mitochondria to cells with non-functional mitochondria has been demonstrated previously.20
Wang et al.12 proposed that Drp1-mediated mitochondrial fragmentation is essential for necroptosis to occur. In contrast to the latter authors, who used a combination of TNF-α, z-VAD-fmk and a Smac mimetic to trigger necroptosis, we found that TNF-α-triggered cell death is Drp1 independent. In fact, based on our experiments using the specific Drp1 inhibitor, Mdivi-1, it appeared that TNF-α-triggered necroptosis and the concomitant release of mitochondria from dying cells are potentiated by the inhibition of Drp1. These differences may potentially be explained by differences in the stimuli used to trigger cell death; indeed, Smac, the antagonist for inhibitor of apoptosis proteins, is a known pro-apoptotic factor.21 Our results do not, however, rule out a role for mitochondrial fission in necroptosis. Notably, during the preparation of the present manuscript, it was shown that Drp1 is dispensable for RIP3-induced necroptosis, in a model of murine embryonic fibroblasts.22 It will be of interest to further characterize the mechanism of release of mitochondria from cells undergoing necroptosis.
We noted that macrophage uptake of mitochondria released from necroptotic cells triggered the secretion of TNF-α, suggesting a possible feed-forward pro-inflammatory signaling loop. Interfering with this signaling through inhibition of necroptosis with Nec-1 or related compounds may prove useful in diseases characterized by an excessive induction of necroptosis. Notably, RIP1 and RIP3 have been reported to have a decisive role during the hyperinflammatory phase of TNF-α-induced systemic inflammatory response syndrome23 and RIP1, but not RIP3, was recently shown to be critical for chronic inflammation in mice that are deficient for SHP-1.24 Moreover, Günther et al.25 demonstrated necroptosis of Paneth cells in the terminal ileum of patients with Crohn’s disease, suggesting a potential role of necroptosis in the pathogenesis of this disease. The authors also provided evidence that the necroptosis inhibitor, Nec-1, is able to reverse the loss of cells in ileal biopsies incubated ex vivo with TNF-α.25 Targeting necroptosis may therefore constitute a novel option in the treatment of patients with inflammatory bowel disease. The release of intact mitochondria, as shown in the present study, and the extracellular release of mtDAMPs such as mtDNA and formyl peptides,7 could contribute to immune activation in inflammatory diseases (see Figure 6b for a schematic representation of these two, mutually non-exclusive scenarios, that is, the release of mtDAMPs versus release of intact mitochondria that act as bona fide danger signals). In conclusion, our data imply that intact mitochondria released during programmed necrosis serve as a novel conduit for signaling between dying cells and the immune system.
Materials and Methods
Reagents
Recombinant human TNF-α was purchased from Gibco/Life Technologies (Carlsbad, CA, USA). The inhibitors, Necrostatin-1 (Nec-1), N-Benzyloxycarbonyl-Val-Ala-Asp (O-Me) fluoromethyl ketone (zVAD-fmk) and 3-(2,4-Dichloro-5-methoxyphenyl)-2,3-dihydro-2-thioxo-4(1H)-quinazolinone (Mdivi-1) were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Cell lines
The parental human Jurkat T-leukemia cells and FADD-deficient Jurkat cells26 kindly provided by Professor John Blenis, Harvard University, were maintained in a humidified 5% CO2 atmosphere in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomysin (Gibco, Paisley, UK). The L929 mouse fibroblast cell line was purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA) and cultured in DMEM medium (Invitrogen/Life Technologies, Carlsbad, CA, USA) supplemented with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified 5% CO2 atmosphere.
Primary immune cells
Human peripheral blood mononuclear cells (PBMCs) from healthy blood donors were separated from buffy coats (Karolinska University Hospital Blood Bank, Stockholm, Sweden) as described before.27 To generate activated, human HMDMs, PBMCs were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin in the presence of 50 ng/ml recombinant macrophage colony-stimulating factor (M-CSF) (R&D Systems, Minneapolis, MN, USA) for 3 days in a humidified 5% CO2 atmosphere. To generate immature, human MDDCs, PBMCs were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin in the presence of 550 IU/ml recombinant human granulocyte-M-CSF (R&D Systems) and 800 IU/ml recombinant human IL-4 (Gibco) at a density of 0.4 × 106 cell/ml in a humidified 10% CO2 atmosphere. After 3 days, the medium with cytokines was replenished and the cells were cultured for 3 days. Peripheral blood neutrophils were isolated from buffy coats from healthy adult blood donors by density gradient centrifugation followed by hypotonic lysis of residual erythrocytes, as described.28 Cells were cultured in RPMI-1640 medium (Sigma-Aldrich), supplemented with 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin.
Annexin V/PI staining
PS exposure was determined using the annexin V-FITC/PI kit (Calbiochem, San Diego, CA, USA). Cells were detected and analyzed with a FACScan flow cytometer (BD Biosciences, San José, CA, USA) equipped with a 488-nm argon laser. Ten thousand events were collected and analyzed using the CellQuest software (BD Biosciences). Cell debris was gated out before analysis based on light-scattering properties.
Plasma membrane disruption
Living cells, as defined by exclusion of trypan blue (Gibco), were counted under a light microscope at × 40 magnification, and those cells that stained blue were scored as membrane-disrupted cells. Cells (200–300) were counted for each sample.
TEM
Non-treated and TNF-α-treated FADD-deficient Jurkat cells were fixed and air-dried onto a carbon film-coated TEM grid, and analysis of morphology was performed using JEM-2100 (JEOL Ltd., Tokyo, Japan) at 200 kV acceleration. For sorted mitochondria, cell culture supernatant after TNF-α treatment was collected and centrifuged at 13 000 × g. The pellet was suspended in mitochondria suspension buffer (MRB; 210 mM mannitol, 10 mM hepes and 1 mM EGTA) and stained with MitoTracker Green FM (Molecular Probes/Life Technologies, Eugene, OR, USA). Thereafter, fluorescence-labeled mitochondria were sorted using a high-speed sorter MoFlo XDP (Beckman Coulter, Fullerton, CA, USA) and ∼3000 MitoTracker-positive events were collected. Sorted mitochondria were fixed and analyzed with TEM as above.
Time-lapse imaging analysis
FADD-deficient Jurkat cells were seeded onto an 18-mm cover glass in a 12-well tissue culture plate. Cells were loaded with fluorescence probes (150 nM MitoTracker Green and 3 μM propiduim iodide, Sigma-Aldrich) in the presence of 10 ng/ml TNF-α in complete medium for 3 h before time-lapse imaging. Time-lapse imaging was performed on a Deltavision Fluorescence microscope (Applied Precision, Issaquah, WA, USA) equipped with a 37 °C climate chamber with an Olympus (Münster, Germany) × 40 Plan Apo 1.4 oil immersion objective. Images of GREEN were acquired with FITC excitation (490/20) and emission (528/38) filters, and of RED with CY3 excitation (555/28) and emission (617/73) filters. The time interval of imaging was 5 min for a total duration of 6 h starting 3 h after TNF-α treatment.
Purification of mitochondria
For purification of released mitochondria, the supernatant from TNF-α-treated, FADD-deficient Jurkat cells were collected and cells and cell debris were eliminated by centrifugation at 700 × g followed by pelleting of the small vesicles at 13 000 × g. The pellet suspended in MRB buffer was separated and centrifuged with percoll gradient at 95 000 × g. The mitochondria band was collected in MRB buffer and designated as ‘mito-pure’. Mitochondria from non-treated cells were isolated using the Mitochondria Isolation Kit (Thermo Scientific, Rockford, IL, USA). The pellet was suspended in MTB buffer and mitochondria were purified by gradient centrifugation as above.
Extraction of mtDNA
Mitochondria were purified from FADD-deficient Jurkat cells as described above. For extraction of mtDNA, the phenol:chloroform:IAA extraction method was used. Briefly, one equal volume of phenol:chloroform:IAA was added to the DNA solution, samples were mixed gently and centrifuged for 5 min at 14 000 × g. Then, the top layer was transferred to a new tube, two volumes of ice-cold 100% ethanol were added, and samples were mixed and kept at −80 °C for 30 min. Samples were then centrifuged at 14 000 × g for 30 min at 4 °C, supernatants were carefully removed, and the pellet (mtDNA) was washed with 70% ethanol and re-suspended in TE buffer. The DNA concentration was determined by a spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Western blotting
Equal amount of supernatant from the pellet of cell culture medium were loaded onto 4–12% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride membranes (Bio-Rad laboratories, Hercules, CA, USA). The membranes were incubated with blocking buffer containing 0.1% Tween-20 and 5% skimmed milk and probed with mouse anti-COX-IV antibody (Molecular Probes) or β-actin antibody (Sigma-Aldrich) as a loading control. Following incubation with horseradish peroxidase-conjugated anti-mouse secondary antibody (DAKOCytomation, Glostrup, Denmark), the bound antibody was visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Phagocytosis assay
Macrophage engulfment of mitochondria was quantified using flow cytometry. Purified released mitochondria were stained with 150 nM MitoTracker Green FM and added to HMDMs in 24-well tissue culture plates in Hank’s Balanced Salt Solution (HBSS) (10 μg/ml mito-pure). Following co-incubation for 1 h, non-engulfed mitochondria were washed off and HMDMs were collected in PBS. Cells were measured on a FACScan flow cytometer (BD Biosciences) and 10 000 events were collected and analyzed using the CellQuest software. Data are reported as % phagocytes positive for uptake of fluorescent targets.
Confocal microscopy
To assess internalization of mitochondria, HMDMs or MDDCs were fixed with 4% paraformaldehyde and mounted on glass slides. Samples were observed under ZEISS LSM510META confocal microscope (ZEISS, Göttingen, Germany) equipped with a × 63 oil immersion objective and data were analyzed using LSM Meta software. Images of green were acquired with FITC excitation (490 nm), of red with CY3 excitation (555 nm), of blue with DAPI excitation (405 nm) and deep red with excitation (633 nm).
Cytokine measurements
Cell culture supernatants were harvested after 1 h co-incubation of HMDMs with mito-pure followed by 5 h incubation in fresh medium, and cells and cell debris were cleared by centrifugation at 13 000 × g. Nine cytokines (TNF-α, IL-1β, IL-6, IL-12p40, IL-18, IFN-α2, IL-4, IL-8, and IL-10) were measured and analyzed with Bio-Plex x-Plex assay kit (Bio-Rad). Data analysis was performed using Bio-Plex Manager software (Bio-Rad).28 Cytokine concentrations were interpolated from an appropriate standard curve. In all analyses, an internal control was incorporated to evaluate interpolate reproducibility. The data shown are from three independent experiments using HMDMs from three donors. To study the effect of purified mtDNA on cytokine production, HMDMs were incubated for 1 h with mtDNA (1, 10, 100, 1000, 10 000 ng/ml) alone or in combination with 100 ng/ml LPS. Neutrophils were first primed with 10 nM fMLP (Sigma-Aldrich) for 2 h and then incubated with mtDNA (1, 10, 100, 1000, 10 000 ng/ml) for 1 h. Cell culture supernatants were collected and stored at −80 °C for further analysis. IL-8 and TNF-α levels were measured by using an ELISA from Invitrogen and Mabtech (Nacka Strand, Sweden), respectively. For TNF-α, the plates were first blocked with 200 μl incubation buffer containing 2% BSA (Sigma-Aldrich) for 1 h at RT.
Detection of surface markers on MDDCs
MDDCs were co-incubated with purified released mitochondria or LPS (100 ng/ml) for 24 h in complete medium. Non-engulfed mitochondria were washed off, and MDCCs were collected in PBS and blocked with PBS containing 1% BSA (Sigma-Aldrich). Cells were labeled with anti-CD80, CD83 or CD86 antibody conjugated with FITC (BD Biosciences) for 30 min. Fluorescence-labeled cells were detected using a FACScan flow cytometer and data were analyzed using the CellQuest software.
Real-time PCR
Mito-pure (purified as described above) were incubated in HBSS (Gibco) and samples were prepared by centrifugation at 13 000 × g for 10 min. MRB buffer only (in HBSS) was included as a negative control. RT-PCR of the mitochondrial Co1 gene was performed according to standard protocols using the Power SYBR Green PCR Master Mix (Applied Biosystems/Life Technologies, Foster City, CA, USA). The primers used were: 5′-ttcgccgaccgttgactattctct-3′ and reverse primer, 5′-aagattattacaaatgcatgggc-3′. To generate a standard curve for quantification of mtDNA in supernatants of the mito-pure samples, mtDNA was extracted from mito-pure using the phenol/chloroform method. A defined amount of purified mtDNA was used as a positive control.
Abbreviations
- COX-IV:
-
cytochrome c oxidase-IV
- DAMPs:
-
damage-associated molecular patterns
- Drp1:
-
dynamin-related protein 1
- FADD:
-
Fas-associated protein with death domain
- HMDM:
-
human monocyte-derived macrophages
- MDDC:
-
monocyte-derived dendritic cells
- PAMPs:
-
pathogen-associated molecular patterns
- RIP:
-
receptor-interacting protein
- TNF:
-
tumor necrosis factor
- HMGB1:
-
high-mobility group box-1
- IAPs:
-
inhibitor of apoptosis proteins
References
Savill J, Dransfield I, Gregory C, Haslett C . A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol 2002; 2: 965–975.
Fadeel B, Xue D . The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit Rev Biochem Mol Biol 2009; 44: 264–277.
Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 2005; 1: 112–119.
Degterev A, Hitomi J, Germscheid M, Ch'en I, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227.
Kaczmarek A, Vandenabeele P, Krysko D . Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 2013; 38: 209–223.
Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010; 464: 104–107.
Collins LV, Hajizadeh S, Holme E, Jonsson IM, Tarkowski A . Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol 2004; 75: 995–1000.
Iyer S, Pulskens W, Sadler J, Butter L, Teske G, Ulland T et al. Necrotic cells trigger a sterile inflammatory response through the Nlrp3 inflammasome. Proc Natl Acad Sci USA 2009; 106: 20388–20393.
Masters S, Walsh P . Release of the mitochondrial endosymbiont helps explain sterile inflammation. Proc Natl Acad Sci USA 2010; 107: E32.
Zhang D-W, Shao J, Lin J, Zhang N, Lu B-J, Lin S-C et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336.
Wang Z, Jiang H, Chen S, Du F, Wang X . The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012; 148: 228–243.
Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 2008; 14: 193–204.
Gray M, Burger G, Lang B . Mitochondrial evolution. Science 1999; 283: 1476–1481.
Underhill D, Goodridge H . Information processing during phagocytosis. Nat Rev Immunol 2012; 12: 492–502.
Welz P-S, Wullaert A, Vlantis K, Kondylis V, Fernández-Majada V, Ermolaeva M et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 2011; 477: 330–334.
Sun S, Sursal T, Adibnia Y, Zhao C, Zheng Y, Li H et al. Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One 2013; 8: e59989.
Scaffidi P, Misteli T, Bianchi ME . Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002; 418: 191–195.
Wickman G, Julian L, Mardilovich K, Schumacher S, Munro J, Rath N et al. Blebs produced by actin-myosin contraction during apoptosis release damage-associated molecular pattern proteins before secondary necrosis occurs. Cell Death Differ 2013; 20: 1293–1305.
Spees JL, Olson SD, Whitney MJ, Prockop DJ . Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci USA 2006; 103: 1283–1238.
Li L, Thomas R, Suzuki H, De Brabander J, Wang X, Harran P . A small molecule Smac mimic potentiates TRAIL- and TNFα-mediated cell death. Science 2004; 305: 1471–1474.
Moujalled DM, Cook WD, Murphy JM, Vaux DL . Necroptosis induced by RIPK3 requires MLKL but not Drp1. Cell Death Dis 2014; 5: e1086.
Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011; 35: 908–918.
Lukens J, Vogel P, Johnson G, Kelliher M, Iwakura Y, Lamkanfi M et al. RIP1-driven autoinflammation targets IL-1α independently of inflammasomes and RIP3. Nature 2013; 498: 224–227.
Günther C, Martini E, Wittkopf N, Amann K, Weigmann B, Neumann H et al. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 2011; 477: 335–339.
Juo P, Woo M, Kuo C, Signorelli P, Biemann H, Hannun Y et al. FADD is required for multiple signaling events downstream of the receptor Fas. Cell Growth Differ 1999; 10: 797–804.
Kagan V, Gleiss B, Tyurina Y, Tyurin V, Elenström-Magnusson C, Liu S-X et al. A role for oxidative stress in apoptosis: oxidation and externalization of phosphatidylserine is required for macrophage clearance of cells undergoing Fas-mediated apoptosis. J Immunol 2002; 169: 487–499.
Farrera C, Fadeel B . Macrophage clearance of neutrophil extracellular traps is a silent process. J Immunol 2013; 191: 2647–2656.
Acknowledgements
We thank J Blenis, Harvard University, for providing FADD-deficient Jurkat cells, K Hultenby for expert assistance with TEM analysis, B Wester for assistance with FACS sorting, M Mousavi for measurements of mtDNA effects on macrophages and neutrophils, F Salomons for assistance with confocal imaging, C Farrera for advice on Luminex measurements, and colleagues in the Fadeel laboratory for illuminating discussions. This research was supported by grants to BF from the Swedish Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellà
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Maeda, A., Fadeel, B. Mitochondria released by cells undergoing TNF-α-induced necroptosis act as danger signals. Cell Death Dis 5, e1312 (2014). https://doi.org/10.1038/cddis.2014.277
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.277
This article is cited by
-
Mitochondria as secretory organelles and therapeutic cargos
Experimental & Molecular Medicine (2024)
-
Emerging roles of mitochondria in animal regeneration
Cell Regeneration (2023)
-
Hypertonicity induces mitochondrial extracellular vesicles (MEVs) that activate TNF-α and β-catenin signaling to promote adipocyte dedifferentiation
Stem Cell Research & Therapy (2023)
-
Intracellular DAMPs in Neurodegeneration and Their Role in Clinical Therapeutics
Molecular Neurobiology (2023)
-
Mitochondrial DNA-Sensing Pathogen Recognition Receptors in Systemic Sclerosis-Associated Interstitial Lung Disease: a Review
Current Treatment Options in Rheumatology (2023)