Abstract
The transcription factor glioma-associated oncogene 1 (Gli1) has been recognized as a very important nuclear executor at the distal end of the Hedgehog (Hh) signal pathway, which has crucial roles in regulating many developmental processes, such as pattern formation, differentiation, proliferation, and apoptosis. Overexpression of patched 1 protein and Gli1 or constitutively active Indian Hedgehog (IHh)-parathyroid hormone-related protein signal pathway may lead to musculoskeletal tumorigenesis. However, for chondrosarcoma few studies have paid close attention to the IHh-Gli1 signal transduction cascade and more work needs to be carried out to fully elucidate Gli1 protein functions. Here we show that the IHh signal pathway was activated in chondrosarcoma, and knocking down the expression of Gli1 attenuated the disturbed IHh signal pathway, which not only suppressed cell proliferation and promoted G2/M cell cycle arrest but also enhanced cell apoptosis by downregulating Bcl-2 and Bcl-xl expression. Furthermore, Gli1 downregulation, not cyclopamine, induced autophagy by regulating mTOR phosphorylation, and inhibition of autophagy prevented Gli1 small interfering RNA-mediated cell death. We also demonstrated that extracellular signal-regulated kinase 1/2 activity may mediate these antiproliferative events induced by Gli1 inhibition. These results indicate that Gli1 inhibition could ultimately provide a promising new approach for chondrosarcoma treatment.
Similar content being viewed by others
Main
Chondrosarcoma is the second most frequently occurring primary bone cancer that develops in cartilage cells.1 At present, surgical resection remains the main form and the most successful means of treating chondrosarcomas, but it continues to have poor prognosis because there is very limited role for adjuvant treatments such as proton beam radiation and chemotherapy;2 in addition, it has been shown that, higher the grade of tumors, worse the prognosis for recurrence and metastasis.3 Therefore, there is an extremely urgent need to explore a novel remedy to combat the cancer.
The Hedgehog (Hh) signal transduction pathway has a crucial role in regulating many developmental processes, such as pattern formation, differentiation, proliferation, death of cells and so on.4 Earlier studies on the Hh pathway mostly focused on embryonic development and neural tissue homeostasis.5 There is growing recognition of the role of Hh genes in the formation of bone and cartilage, especially for the Indian Hedgehog (IHh), which is one of the key signaling molecules controlling both chondrocyte hypertrophy and bone formation in the developing skeletal system.6 IHh is secreted by prehypertrophic and hypertrophic chondrocytes in the normal growth plate of metazoans.7 On one side, IHh ligands have been demonstrated to bind to patched 1 protein (PTCH1), causing internalization and degradation, thereby releasing SMO into the primary cilia where it promotes GLI protein dissociation through its inhibitory complex sufu/Gli8, 9 and then activates the glioma-associated (Gli) family of zinc finger transcription factors. GLI-mediated transcriptional activation resulted in the upregulation of target genes including wg (WNT family), dpp (TGFβ/BMP family), and ptc (PTCH.PTCH2,TCR8),10 and stimulated the proliferation and differentiation of cartilage cells.11 On the other hand, IHh activated the expression of parathyroid hormone-related protein (PTHrP) in the periarticular cells and articular chondrocytes. PTHrP signals through its receptor PTHR1 to inhibit chondrocyte hypertrophy and suppresses further IHh expression by keeping chondrocytes in a proliferating state; thus, it can be seen that IHh and PTHrP signaling forms a negative feedback loop that modulates the development of the normal growth plate.12, 13 For chondrosarcoma, the IHh-PTHrP signal pathway is better understood and its function has been studied in great detail, but few studies have paid close attention to the IHh-glioma-associated oncogene 1 (Gli1) signal transduction cascade and more work needs to be carried out to fully elucidate Gli1 protein functions. Current strategies have been focusing on the development and use of small molecules to inhibit smoothened (Smo) (such as Cyclopamine, GDC-0449, LDE225, BMS-833923 (XL139), IPI-926, PF-04449913, LEQ506, and TAK-441).14 However, these strategies might not be applicable to the treatment of tumors that harbor molecular lesions downstream of Smo. In earlier studies, we have illustrated that constitutive activation of the Hh signaling pathway in chondrosarcoma is rarely caused by PTCH1 or SMO mutations,15 and therefore the aberrant activation of the pathway in chondrosarcoma is more likely due to downstream molecules, such as GLI proteins, which act as very important nuclear executors and mediate Hh signaling at the distal end of the pathway. On the basis of recent data on GLI transcription factors we have concluded that GLI1, and not GLI2 or Gli3, is the direct transcriptional Hh-target gene, which determines the cell’s response to the Hh pathway activity and might be the causal agent in multiple cellular processes such as proliferation, cell cycle, and cell survival. In this paper, we investigated the effects of Gli1 small interfering RNA (siRNA) on human chondrosarcoma cell lines SW1353 and JJ012. Our results indicated that knockdown of Gli1 expression not only attenuated the affected Hh signal pathway but also suppressed cell growth and cell cycle progression, and induced apoptosis as well as autophagy, and the extracellular signal-regulated kinases (ERK)1/2 activity may mediate these antiproliferative events induced by Gli1 inhibition. Thus, blocking the expression of Gli1 would be an ideal strategy for combating chondrosarcoma.
Results
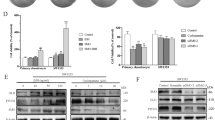
Expression of IHh pathway members in the normal articular cartilage, in chondrosarcoma, and in their corresponding cell lines HC-a, SW1353, and JJ012
We first examined the expressions of key members in the IHh pathway using immunohistochemistry (IHC), qPCR and western blot analysis (WB) in 20 samples taken from the normal articular cartilage and chondrosarcoma tissues. qPCR data are not shown here because the CT values in articular cartilage samples were too high to obtain apart from IHh. IHC (Figure 1a) and WB (Figure 1b) analyses showed that PTHrP, PTCH1, SMO, and Gli1 were all expressed in chondrosarcoma but not in the normal articular cartilage tissues. On the other hand, expression of IHh was detected in both tissues, but was significantly elevated in chondrosarcoma compared with the articular cartilage (Figure 1b). As visualized by immunohistochemistry, Gli1 displayed an absolutely nuclear expression pattern, whereas others stained on cell membranes and in the cytoplasm (Figure 1a). To further validate our results, we analyzed the expression of these genes in their corresponding cell lines HC-a, SW1353, and JJ012 by PCR and WB. Similar results were obtained (Figures 1c and d). These findings indicate that there was an active IHh pathway in chondrosarcoma, which is essential for tumor growth, recurrence, metastasis, and cell survival.16
Expression of IHh pathway members in the normal articular cartilage, in chondrosarcoma. and in their corresponding cell lines HC-a SW1353 and JJ012. (a and b) IHC and WB analyses for IHh, PTHrP, PTCH, SMO, and Gli1 were performed in tissue sections from the normal articular cartilage and chondrosarcoma, and the staining intensities and location of IHC were evaluated. (c and d) Next, these proteins were examined by qPCR and WB in human articular chondrocyte cell line HC-a and chondrosarcoma cell lines SW1353 and JJ012 (*P<0.05, **P<0.01 versus HC-a)
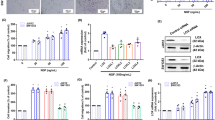
Cell viability was reduced by Gli1 siRNA but not by the SMO inhibitor cyclopamine
As the IHh pathway is activated in chondrosarcoma, it was interesting to study whether inhibition of the pathway with a natural small-molecule cyclopamine, a specific SMO inhibitor, would kill SW1353 and JJ012 cells. Our results showed that adding up to 20 μM cyclopamine in the culture medium did not affect cell viability in the two chondrosarcoma cell lines (Figure 2a). On the basis of the results, the activator of the pathway in chondrosarcoma might be located downstream of SMO, being a transcription factor such as GLI1. To test this hypothesis, we used siRNA to knock down Gli1 expression to see whether Gli1 was involved in the regulation of viability and proliferation in the chondrosarcoma cell lines. Our results showed that the mRNA and protein levels of Gli1 in SW1353 and JJ012 cells were reduced significantly 24, 48, and 72 h after siGli1 transfection; data collected 48 h after the knockdown treatment are shown in Figures 2b and c. The cell viabilities assayed by MTS showed constant decline with increase in time after the Gli1 siRNA transfection and were only about 30–55% of the values before the transfection in JJ012 and SW1352, respectively (Figure 2d).
Cell viability was reduced by Gli1 siRNA but not by the SMO inhibitor cyclopamine. (a and b) The Gli1 siRNA transfection efficiency was measured by qPCR and WB. (c) Cell viability of cyclopamine and Gli1 siRNA in chondrosarcoma cell lines was tested by MTS assay; cells were incubated with 0, 5, 10, and 20 μM of cyclopamine for 24 h (P>0.05 versus control). (d) Cells were treated with Gli1 siRNA for the indicated time. All data are expressed as the mean±S.D. of three independent experiments (*P<0.05, **P<0.01 versus controls)
The active IHh signal pathway can be attenuated by siGli1 but not by cyclopamine
To further verify the findings, we assessed the net effect of siGli1 or cyclopamine on the expression of the key members in the pathway. We found that RNAi-mediated knockdown of Gli1 in chondrosarcoma cell lines diminished not only the secretion of IHh and PTHrP but also the expression of PTCH1 and SMO, whereas cyclopamine did not affect their expression except that of SMO (Figures 3a and b). This is in contrast to previous studies in which cyclopamine clearly downregulated GLI1 and PTCH1.17, 18 That is, the whole Hh signal transduction cascade was weakened by Gli1 siRNA but not by the SMO inhibitor cyclopamine.
The active IHh signal pathway can be attenuated by siGli1 but not by cyclopamine. Cells s were treated with Gli1 siRNA and cyclopamine (10 μM) for 24 h. Changes in the expression of IHh, PTHrP, PTCH, SMO, and Gli1 were examined by qPCR (a) and WB (b). The data are presented as mean±S.D. of three independent experiments (*P<0.05, **P<0.01, #P>0.05 versus controls)
Gli1 inhibition promoted G2/M cell cycle arrest and apoptosis
Forty-eight hours after siGli1 treatment, the cells appeared to stop proliferation and arrested at the G2/M phase (Figure 4a). As a result, the proportion of cells at the G2/M stage was increased by 13.88±0.76% and 13.68±1.16% in SW1353 and JJ012, respectively, compared with untreated controls (Figure 4c). As shown by flow cytometric analysis, there were increased proportions of early apoptotic cells (marked as LR in Figure 4b) and late apoptotic cells (marked as UR in Figure 4b) 48 h after siGli1 transfection, which increased by 14.89±0.82% and 27.69±1.24% in SW1353 and JJ012, respectively, as compared with controls (Figure 4d).
Gli1 inhibition promoted G2/M cell cycle arrest and apoptosis. (a) Cells treated with siGli1 for 48 h were fixed in 70% ethanol, permeabilized in Triton-X, digested with RNAse A, and stained with propidium iodide. (b) Apoptotic cells were measured with the Annexin V/FITC kit according to the manufacturer’s instructions and analyzed by flow cytometry after treatment with siGli1 for 48 h. (c) The bar chart shows the percentage of cells in cell cycle. The treatment groups are labeled on the horizontal axis. The siGli1 treatment led to a significant increase in the proportion of cells in the G2/M phase in the two cell lines tested. (d) The pictures demonstrate one of three similar experiments in chondrosarcoma cell lines and show a significant increase in the early and late apoptotic index
Correlative changes of molecules in the process of proliferation, cell cycle, and apoptosis after treatment with siGli1
To further test the functions of Gli1 in the process of proliferation, cell cycle, and apoptosis we performed immunocytochemical analysis for Ki-67 and WB analysis for proliferating cell nuclear antigen (PCNA), CyclinB1, pRb, Bax, Bcl-2, BCL-xl, and Caspase3. PCNA functions in cell cycle progression and serves as an attractive assay of cell proliferation;19 Ki-67 is a nuclear protein that is expressed in proliferating cells and may be required for maintaining cell proliferation. Ki-67 has been used as a marker for cell proliferation of solid tumors and some hematological malignancies.20 CyclinB has an integral role in the progression of cells into and out of M phase, which is necessary for cells to enter mitosis and therefore necessary for cell division.21 Rb is a tumor suppressor protein that has important roles in the checkpoint of cell cycle. Downregulation of pRb expression increases both the S- and G2/M phase cell fractions of lung fibroblasts.22 In our study, the expressions of PCNA, Ki-67, cyclinB, and pRb were inhibited after treatment with siGli1 for 48 h in chondrosarcoma cell lines (Figure 5). These data supported our in vitro evidence pointing to the critical role of Gli1 in the G2/M mitotic/transition phase.
To understand the mechanism of apoptosis exerted by siGli1, we studied the effect of siGli1 on the expression of pro-apoptotic protein Bax, anti-apoptotic proteins Bcl-2 and Bcl-xl, and caspase3. As shown in Figure 5, there were significant inhibitions of Bcl-2 and Bcl-xl expression after treatment with siGli1 for 48 h, but no obvious changes in the expression of Bax. In addition, cleavage of caspase3, which requires proteolytic processing of its inactive zymogen into activated p17 and p12 fragments,23 had not been detected during the execution of the apoptotic cascade (Figure 5). These observations suggest that the Bcl-2 family of proteins, but not caspase3, controls apoptotic programmed cell death.
Gli1 downregulation, not cyclopamine, induced autophagy by regulating mTOR phosphorylation
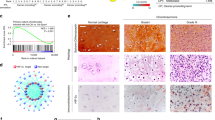
Autophagy-associated cell death is another type of programmed cell death after the discovery of apoptosis and necrosis by maintaining an important homeostatic mechanism that removes damaged or unnecessary proteins and organelles and recycles cytoplasmic contents through a process by which these components are engulfed in autophagosomes and then digested via the lysosomal pathway.24 Induction of autophagy might be a novel approach for anticancer therapy. To investigate whether Gli1 is involved in autophagy, transmission electron microscopy was performed to examine the ultrastructures of autophagy. Representative images depicting the ultrastructures of autophagy, such as autophagosome-like vacuoles with double-membrane structures (Figures 6a and b black arrow) and segments of the double-membrane formed between a vacuole and mitochondrion (Figures 6a and c white arrow), are shown in Figure 6a. Expression of LC3-II visualized by immunofluorescence is recognized as a means to track changes in autophagosome formation in living cells. LC3 is processed from LC3-I to LC3-II during autophagy. LC3-II is inserted into newly formed autophagosome membranes.25 Therefore, we investigated the impact of Gli1 expression on LC3-II production. Our results show that silencing Gli1 with siRNAs increased the LC3-II localization in the cytoplasm in both cell lines (Figure 6b) and resulted in the accumulation of LC3-II and degradation of P62 (a known autophagy substrate) (Figure 6c). In contrast, no such effect was detected in cyclopamine-treated cells (Figure 6d).
Gli1 downregulation, not cyclopamine, induced autophagy by regulating mTOR phosphorylation. (a) Representative TEM images depict ultrastructures of autophagy SW1353 and JJ012 cells transfected with Gli1 siRNA for 24 h. Autophagosome-like vacuoles with double-membrane structures (black arrow) and segment of the double-membrane being formed between a vacuole and mitochondrion (white arrow). (b) Cells were grown on coverslips and transfected with Gli1 siRNA for 24 h. They were then incubated overnight at 4 °C with rabbit polyclonal anti-LC3 Ab (1 : 200) and then sequentially in anti-rabbit IgG conjugated with Dylight 488 (1 : 400) for 2 h at room temperature. The cells were then visualized by confocal microscopy. The data were normally distributed and statistically analyzed using one-way ANOVA. The double asterisk denotes that the group is statistically different from the control groups (**P<0.01). (c) WB was used to detect the expression of LC3-II (an autophagosome marker) and P62 (a known autophagy substrate) after transfection with Gli1 siRNA and being incubated with cyclopamine (10 μM) for 24 h. Downregulated expression of Gli1 induced accumulation of Lc3-II and degradation of p62 (a known autophagy substrate). (d) However, cyclopamine had no effect on Lc3-II and p62. (e) The relationship between Gli1 and mTOR was also studied. (f) Densitometry was performed for quantification, and the ratios of P-mTOR and P-p70S6K to β-actin are presented
To further explore the contribution of Gli1 to autophagy regulation, we studied the relationship between Gli1 and mTOR. We observed that silenced expression of Gli1 decreased mTOR phosphorylation, but rapamycin, which stimulates autophagy through mechanisms involving mTOR kinase inactivation, had no effect on Gli1. We also analyzed the phosphorylation of p70S6K, a marker of mTOR activity, and found that it followed the same trend as mTOR (Figure 6e). Densitometry was performed for quantification, and the ratios of P-mTOR and P- p70S6K to β-actin are presented in Figure 6f. These outcomes elicited downregulated expression of Gli1-induced autophagy depending on mTOR dephosphorylation.
Autophagy inhibition prevented siGli1-mediated cell death
Autophagy has been associated with both cell death and survival depending on the cellular context and stimulus.26 Given that autophagy can be induced by siGli1, we evaluated whether autophagy induced by Gli1 inhibition had a role in cell death. A link between autophagy and cell death has been demonstrated using pharmacological (e.g., 3-Methyladenine (3-MA), chloroquine (CQ), bafilomycin A1, or monensin) and genetic (silencing of Atg5, Atg7, Beclin1, Atg10, or Atg12) approaches for suppression of autophagy.27 3-MA can block the induction of LC3-II puncta formation.28 CQ is a lysosomotropic agent that has been suggested to inhibit autophagy by perturbing lysosomal function.27 Cells treated with siGli1 in the presence of 3-MA (2.5 mM) and CQ (30 μg/ml) demonstrated that 3-MA and CQ significantly enhanced the viability of cells in response to Gli1 inhibition (Figure 7a). To further test this possibility, we invalidated the expression of autophagic genes Atg7 and Beclin1 (Figure 7b) and quantified the production of LC3-II (Figure 7c). Similarly, RNA interference (RNAi) directed against the autophagy genes atg7 and beclin1 blocked cell death in chondrosarcoma cells transfected with siGli1 (Figure 7d). These results support the possibility that autophagy inhibition prevented siGli1-mediated cell death.
Autophagy inhibition prevented siGli1-mediated cell death. (a) Cells were transfected with Gli1 siRNA. After 6 h, cells were exposed to 3-MA (2.5 mM) or CQ (30 μg/ml) for 48 h. Then cell viability was tested using an MTS assay. (b and c) SW1353 and JJ012 cells were transfected with siGli1 in the presence of Atg7 and Beclin1 siRNA; 48 h after transfection, the cells were analyzed by immunoblotting with the indicated antibodies. (d) The viability of SW1353 and JJ012 cells in (c) was analyzed by the MTS assay as described in Materials and Methods. Data are presented as mean±S.D. and are representative of three independent experiments (*P<0.05, **P<0.01 versus controls)
ERK1/2 activity mediated antiproliferative events
The ERK signaling pathway has a crucial role in almost all cell functions and therefore requires exquisite control of its spatiotemporal activity.29 On treatment of Gli1 siRNA-transfected cells with the MAPK/ERK kinase inhibitor U0126 (10 μM; U0126 is not toxic up to 10 μM in SW1353 and JJ012 cells) we found that U0126 improved the cell vitality that had been reduced by Gli1 siRNA (Figure 8a), resulting in the release of most cells from arrest at the G2/M stage (Figure 8b) and also strongly reduced apoptosis (Figure 8c) in Gli1 siRNA-transfected chondrosarcoma cells. In addition, U0126 also significantly reduced LC3-II puncta formation, which implied that Gli1 inhibition-induced autophagy was attenuated in U0126 treatment (Figure 8d), and the MEK/ERK-related signaling pathway contributed to autophagy in the chondrosarcoma cell lines. To further determine whether the involvement of MEK/ERK signaling is a direct or indirect consequence of depressed Gli1 activity, we treated the cells with siGli1 and U0126 for 24 h and found that reduction in Gli1 expression promoted ERK phosphorylation but had no impact on MEK1/2 and that U0126 inhibited both activated and inactivated MEK1/2 as well as activated ERK1/2 pathways, leading to the repression of GLI1. Taken together, the current data showing that depressed Gli1 activity promoted ERK activity and that Gli1 activity was also dependent on MEK/ERK signaling highlight the possible presence of a novel feedback mechanism that modulates GLI1 activity (Figure 8e). Many studies associate the MEK/ERK signal pathway’s oncogenic potential to increased cell survival. Paradoxically, depending on the cell type and stimulus, ERK activity has been shown to mediate different antiproliferative events such as apoptosis, autophagy, and senescence in vitro and in vivo.30 Therefore, we hold the opinion that phosphorylation-ERK contributed to the cell cycle arrest, apoptosis, and autophagy induced by Gli1 inhibition in chondrosarcoma cell lines.
ERK1/2 activity mediated antiproliferative events. Cells were transfected with Gli1 siRNA. After 6 h the cells were exposed to the MAPK/ERK kinase inhibitor U0126 (10 μM) or not for 24 h. (a) Cell viability was tested using an MTS assay. (b and c) Cell cycle and apoptosis were analyzed by flow cytometry. (d) LC3-II puncta formation was visualized by confocal microscopy. (e) Thereafter, cells were exposed to siGli1 and U0126 for 24 h to determine whether the involvement of MEK/ERK signaling is a direct or indirect consequence of depressed Gli1 activity. Cell lysates were prepared and analyzed by immunoblotting using the indicated antibodies (*P<0.05, **P<0.01 versus controls). (f) Detailed molecular regulation mechansim of Gli1. (→) Stimulatory modification. (⊥) Inhibitory modification. (X) Signal dysfunction
Discussion
According to our studies, we find that the IHh signal pathway is disturbed in chondrosarcoma. Overexpression of PTCH1 and Gli1 or a constitutively active IHh-PTHrp signal pathway may lead to musculoskeletal tumorigenesis.31, 32, 33 Coincidentally, it is exciting to discover that knockdown of Gli1 expression by siRNA downregulated the expression of major Hh signal pathway members, including IHh, PTHrP, PTCH1, and SMO. This was probably due to insufficient Gli1 protein translocation into the nucleus to initiate the transcription of the Hh pathway genes, such as Gli1 and PTCH1, as well as many survival-promoting molecules.14 In the present study, SMO levels appeared to be insufficient for stimulating the pathway,34 so did chondrosarcoma in our study, but the expression of SMO was downregulated by siGli1. Thus, the whole Hh signal transduction cascade was weakened and corrected by knockdown of Gli1 expression. We also confirmed that siGli1 decreased the proliferation rate as well as PCNA and Ki-67 expression, which has already been abundantly implicated in other cancers, such as in BCCs and meduloblastomas.35, 36 Recent work has provided insight into the fact that Hh influences the expression and activity of core cell cycle components at multiple points within the cell cycle in a variety of circumstances.37 Intriguingly, we found that siGli1 induced cell cycle G2/M phase arrest by downregulating cyclinB and Rb phosphorylation, thereby rendering chondrosarcoma cells quiescent and prohibiting tumor progression. Furthermore, the Bcl-2 protein family has a central role in the control of apoptosis and is also thought to be involved in resistance to conventional cancer treatment.38 It is the Gli1 siRNA that affected Bcl-2 and Bcl-xl expression and led to apoptosis in sw1353 and JJ012 cells.
Autophagy is an important homeostatic mechanism that removes damaged or unnecessary proteins and dysfunctional cellular organelles and recycles cytoplasmic contents in all living cells.39 The process of autophagy was initially identified as a cell survival mechanism in response to stress.40 However, under certain conditions, autophagy results in a form of cell death now described as type II programmed cell death.41 It had been reported that the Hh signaling pathway regulates autophagy.42, 43, 44 Jimenez-Sanchez et al.42 have confirmed that Gli2 is necessary for the inhibition of autophagy by Hh signal, but they were unable to completely exclude the contribution of Gli1 and Gli3 to autophagy regulation. The mechanism by which Gli1 regulates autophagy is still unclear and the role of autophagy in cell growth and cell death in chondrosarcoma is still being debated considerably. In our study, it was found, for the first time to our knowledge, that Gli1 inhibition, not cyclopamine, induced synthesis of autophagosome in chondrosarcoma. Autophagy has been implicated in maintaining cancer cell survival, but its precise role is ambiguous. Autophagy also functions as a mechanism of pro-cell death, depending on the cellular context that when unrestrained can result in progressive consumption of cellular components and subsequent cell death and might mediate resistance to anticancer therapies such as radiation, chemotherapy, and some targeted therapies.45, 46, 47 In addition, autophagy inhibition prevented siGli1-mediated cell death. It strengthened our point of view that induction of autophagic cell death is a useful alternative approach to killing tumor cells. Induction of autophagy is a possible antitumor mechanism of action of allosteric inhibitors of mTORC1. One such rapalog, rapamycin, is used as an antirejection drug in organ transplant recipients and has demonstrated anticancer activity in myxoid chondrosarcoma as well.48 Recent studies have shown that activated mTOR/S6K1 pathway promotes Gli1 transcriptional activity and oncogenic function,49 but our study hold the opposite view that Gli1 transcription factor appears to be no changes treated with rapamycin, whereas silenced the expression of Gli1 with siRNAs lead to mTOR dephosphorylation then induced autophagy-associated cell death suggested that Gli1 may be located at the upstream of mTOR in chondrosarcoma cells. Consequently, when treating resistant tumors such as recurrent chondrosarcoma, antineoplastics leading to autophagy-associated cell death might prove to be a unique form of combinational therapy rather than a novel target for anticancer therapy.46
In the investigations on the mechanism of these antiproliferative events induced by Gli1 inhibition, it has been shown that constitutive activation of ERK by active Raf,50 cadmium,51 or IGF-I receptor52 induces a form of cell death that is correlated with extensive cell rounding and with the formation of cytoplasmic macrovacuoles, which pushes the nucleus and the cytoplasm to the side of the quiescent and dying cell. We also found that there is a crosstalk between the Hh and the MEK/ERK signal pathway and that ERK1/2 activity mediates these antiproliferative events. On the basis of these findings, we propose a tentative regulatory mechansim of IHh and Gli1 in Figure 8f.
However, it should be noted that the study we had examined is limited to in vitro cell lines, and a significant number of challenges still need to be overcome for a successful in vivo study. Although there are animal studies that do support the possibility of an RNAi mechanism of action from administered siRNA,53 our in vivo study, using naked siRNA or siRNA by liposomal delivery, suggested non-RNAi mechanisms of action for chondrosarcoma suppression. Therefore, we are trying to find a new delivery system that can provide effective delivery of functional siRNA to animals with chondrosarcoma.
In conclusion, knockdown of IHh-Gli1 signaling by Gli1-specific siRNAs not only attenuated the disturbed IHh signal pathway but also curbed the growth and survival of the treated chondrosarcoma cells. The related mechanisms were also systematically demonstrated in this paper. This will not only advance our understanding of chondrosarcoma development at the molecular level but also lead to an important new opportunity in which therapeutic effects might be achieved by Gli1 inhibitors.
Materials and Methods
Human articular cartilage and chondrosarcoma specimens, cell lines, and reagents
The fresh human normal articular cartilage and chondrosarcoma specimen were obtained under the approved protocols of the Institutional Ethics Review Boards of Peking University People’s Hospital. Informed consent for the experimental use of surgical specimens was obtained from all patients in written form according to the hospital’s ethical guidelines. Excisions obtained after surgery were properly conserved for the experiment. The human articular chondrocyte cell line HC-a (Sciencell, Carlsbad, CA, USA) and two chondrosarcoma cell lines SW1353 (ATCC HTB-94; Manassas, VA, USA) and JJ012 (JJ012; a gift from Dr. J Block, Rush Medical College, Chicago, IL, USA) were used in the study. HC-a cells were maintained in DMEM supplemented with 10% fetal bovine serum, plus antibiotics. SW1353 was maintained in L-15 medium, whereas JJ012 cells were grown in RPMI-1640 medium supplemented with FBS and antibiotics. The following reagents were purchased from Sigma-Aldrich (Sciencell): cyclopamine (C4116), 3-Methyladenine (M9281), 3-Methyladenine U0126 (U120), and polyclonal antibodies against LC3 (L7543) and P62 (N1163). Rabbit monoclonal anti-IHh (ab39634), anti-PTHrP (ab16827), anti-PT CHed (ab39266), and anti-SMO (ab5694) were purchased from Abcam (Abcam, Cambridge, UK). Anti-Gli1 (3538), anti-Phospho-MEK1/2 (9154), anti-MEK1/2 (9126), anti-Phospho-ERK1/2 (4344), anti-ERK1/2 (4695), and anti-β-actin were from Cell Signaling Technology (Beverly, MA, USA).
Immunohistochemistry
Paraffin sections were reacted with rabbit polyclonal anti-IHh, anti-PTHrP, anti-PTCH, anti-SMO, and anti-Gli1 antibodies (1 : 200 dilution). Sections stained with nonimmune rabbit serum (1 : 200 dilution) in phosphate-buffered saline (PBS) instead of primary antibody served as negative controls. Cells exhibiting positive staining on cell membranes and in the cytoplasm and nucleus were counted in at least 10 representative fields ( × 400 magnification) and the mean percentage of positive cells was calculated. Immunostaining was assessed by two independent pathologists blinded to clinical characteristics and outcomes.
RNAi and cell transfection
The siRNAs against Gli1 were designed by OriGene Technologies (Rockville, MD, USA, SR301820). The siRNAs, 21nt-long, double-stranded, were purified and desalted by the provider. Their sequences were as follows: sense strand siGli1 primer 5′-GCCACCAAGCUAACCUCAUGUTT-3′ and antisense strand siGli1 primer 5′-ACAUGAGGUUAGCUUGGUGGCTT-3′. The following nonspecific siRNAs were used as control: forward 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse 5′-ACGUGACACGUUCGGAGAATT-3′. For transfection, cells were transfected with the siRNAs using the Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions after cells were grown to ∼60% confluence.
Cell viability assay
Cell viability was investigated using an MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (inner salt; Sigma, M2003) assay. The cells were suspended in 0.2 ml of medium at a concentration of 5000 cells/well and incubated overnight in 96-well plates. Absorbance at 490 nm was measured using a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Cell cycle and apoptosis analysis by flow cytometry
Cells were fixed in 70% ethanol, permeabilized in Triton-X, digested with RNAse A, and stained with propidium iodide. Apoptotic cells were measured with Annexin V/FITC kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions and analyzed by flow cytometry after compound treatment as described previously.54
RNA extraction and qPCR analysis
Total RNA was isolated from SW1353 and JJ012 cells with Trizol reagent (Oakville, Ontario, Canada), according to the manufacturer’s instructions. After purification, RNA was converted to complementary DNA (cDNA) using the First-Strand cDNA synthesis kit (ReverTra Ace kit Toyobo, Tokyo, Japan) according to the manufacturer’s instructions. The cDNA was amplified from 2 μg of total RNA using Super Script one-step RT-PCR or qPCR with the platinum Taq system (Invitrogen). The primers used for amplification of IHh, PTHrP, PTCH,SMO, Gli1, and GAPDH transcripts were as follows: IHh forward 5′-CGTCGTGGTGTAGTCATAG-3′ and IHh reverse 5′-GGATGTGTCTTGAGGAGTG-3′; PTHrP forward 5′-CGATTCTTCCTTCACCATCT-3′ and PTHrP reverse 5′-CGTCTCCACCTTGTTAGTT-3′; PTCH forward 5′-TGTGATGGATGCTGGAACCT-3′ and PTCH reverse 5-′CCTACATAATGCCTGCCTCTAC-3′; SMO forward 5′-GTTCTCCTATATCCTCCTTCTC-3′ and SMO reverse 5′-CTTCCTGCTGTCTGATTCTT-3′; Gli1 forward 5′-CCAACCTCTGTCTACTCAC-3′ and Gli1 reverse 5′-CCTGTTCTGGCTTGACTT-3′; GAPDH forward 5-′GAAGGTCGGAGTCAACGGATTT-3′ and GAPDH reverse 5-′ATGGGTGGAATC;ATATTGGAAC-3′. The reactions were performed in a 96-well optical plate (Applied Biosystems, Warrington, UK) at 94 °C for 2 min, followed by 38 cycles of 94 °C for 45 s, 56 °C for 45 s, and 72 °C for 40 s.
Western blotting and Immunofluorescence analysis
For western blotting, equal amounts of proteins collected from different cell lysates or fractions were resolved on 8–15% SDS-PAGE using a NuPAGE system (Invitrogen), and then transferred onto PVDF membranes according to the manufacturer’s instructions.55 For immunofluorescence, cells were grown on coverslips and transfected with the Gli1 siRNA, incubated overnight at 4 °C with rabbit polyclonal anti-LC3 Ab (1 : 200), and then reacted with anti-rabbit IgG conjugated with Dylight 488 (1 : 400) for 2 h at room temperature. After washing with PBS, the cells were mounted on vectashield and visualized using confocal microscopy (Leica, Heidelberg, Germany).56, 57
Transmission electron microscopy
Transmission Electron Microscopy (TEM) was performed on cells 48 h after Gli1 siRNA transfection. Cells were digested with 0.25% trypsin and suspended at a density of 1.0 × 106/ml. Then 3% glutaraldehyde was added for 3 h at 4 °C for fixation. Ultrathin sections (100 nm) were prepared, stained with uranyl acetate and lead citrate, and examined under an electron transmission microscope (H-600; Hitachi, Tokyo, Japan).
Statistical analysis
The SPSS19.0 software package was used to perform all statistical analyses. Data were analyzed by one-way analysis of variance with the multiple comparison test of Bonferroni. Unpaired Student’s t-tests were used for comparison between two groups. Data are shown as mean±S.D. A value of P<0.05 was considered statistically significant.
Abbreviations
- siNC:
-
small interfering RNA negative control
- siGli1:
-
Gli1 small interfering RNA
- IHh:
-
Indian Hedgehog
- PTHrP:
-
parathyroid hormone-related protein
- PTCH1:
-
patched 1 protein
- Smo:
-
smoothened
- Gli1:
-
glioma-associated oncogene 1
- Erk:
-
extracellular signal-regulated kinases
References
O'Neal LW, Ackerman LV . Chondrosarcoma of bone. Cancer 1952; 5: 551–577.
Lee FY, Mankin HJ, Fondren G, Gebhardt MC, Springfield DS, Rosenberg AE et al. chondrosarcoma of bone: an assessment of outcome*. J Bone Joint Surg 1999; 81: 326–338.
Fiorenza F, Abudu A, Grimer R, Carter S, Tillman R, Ayoub K et al. Risk factors for survival and local control in chondrosarcoma of bone. J Bone Joint Surg, Br 2002; 84: 93–99.
Nybakken K, Perrimon N . Hedgehog signal transduction: recent findings. Curr Opin Genet Dev 2002; 12: 503–511.
Ingham PW, McMahon AP . Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–3087.
Mak KK, Kronenberg HM, Chuang P-T, Mackem S, Yang Y . Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Sci Signaling 2008; 135: 1947.
Kobayashi T, Chung U-i, Schipani E, Starbuck M, Karsenty G, Katagiri T et al. PTHrP and Indian hedgehog control differentiation of growth plate chondrocytes at multiple steps. Development 2002; 129: 2977–2986.
Huangfu D, Anderson KV . Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 2006; 133: 3–14.
Jia J, Jiang J . Decoding the Hedgehog signal in animal development. Cell Mol Life Sci 2006; 63: 1249–1265.
Bale AE . Hedgehog signaling and human disease. Annu Rev Genomics Hum Genet 2002; 3: 47–65.
Lum L, Beachy PA . The Hedgehog response network: sensors, switches, and routers. Science 2004; 304: 1755–1759.
Lanske B, Karaplis AC, Lee K, Luz A, Vortkamp A, Pirro A et al. PTH/PTHrP receptor in early development and indian hedgehog—regulated bone growth. Science 1996; 273: 663–666.
Henderson JE, Amizuka N, Warshawsky H, Biasotto D, Lanske B, Goltzman D et al. Nucleolar localization of parathyroid hormone-related peptide enhances survival of chondrocytes under conditions that promote apoptotic cell death. Mol Cell Biol 1995; 15: 4064–4075.
Ng JM, Curran T . The Hedgehog's tale: developing strategies for targeting cancer. Nat Rev Cancer 2011; 11: 493–501.
Yan T, Alman BA, Wunder JS . Patched-one or smoothened gene mutations are infrequent in chondrosarcoma. Clin Orthop Relat Res 2008; 466: 2184–2189.
Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P et al. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med 2009; 1: 338–351.
Clement V, Sanchez P, de Tribolet N, Radovanovic I, i Altaba AR . HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal and tumorigenicity. Curr Biol 2007; 17: 165.
Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA 2007; 104: 5895–5900.
Kubben F, Peeters-Haesevoets A, Engels L, Baeten C, Schutte B, Arends J et al. Proliferating cell nuclear antigen (PCNA): a new marker to study human colonic cell proliferation. Gut 1994; 35: 530–535.
Gerdes J, Lemke H, Baisch H, Wacker H-H, Schwab U, Stein H . Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 1984; 133: 1710–1715.
Ford HL, Pardee AB . Cancer and the cell cycle. J Cell Biochem 1999; 75: 166–172.
Giacinti C, Giordano A . RB and cell cycle progression. Oncogene 2006; 25: 5220–5227.
Porter AG, Jänicke RU . Emerging roles of caspase-3 in apoptosis. Cell Death Differ 1999; 6: 99.
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G . Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8: 741–752.
Tanida I, Ueno T, Kominami E . LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004; 36: 2503–2518.
Codogno P, Meijer AJ . Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ 2005; 12: 1509–1518.
Boya P, González-Polo R-A, Casares N, Perfettini J-L, Dessen P, Larochette N et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005; 25: 1025–1040.
Seglen PO, Gordon PB . 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA 1982; 79: 1889–1892.
Kohno M, Pouyssegur J . Targeting the ERK signaling pathway in cancer therapy. Ann Med 2006; 38: 200–211.
Cagnol S, Chambard JC . ERK and cell death: Mechanisms of ERK-induced cell death–apoptosis, autophagy and senescence. FEBS J 2010; 277: 2–21.
Bovée JV, van den Broek LJ, Cleton-Jansen A-M, Hogendoorn PC . Up-regulation of PTHrP and Bcl-2 expression characterizes the progression of osteochondroma towards peripheral chondrosarcoma and is a late event in central chondrosarcoma. Lab Invest 2000; 80: 1925–1933.
Pateder DB, Gish MW, O'Keefe RJ, Hicks DG, Teot LA, Rosier RN . Parathyroid hormone-related peptide expression in cartilaginous tumors. Clin Orthop Relat Res 2002; 403: 198–204.
Tiet TD, Alman BA . Developmental pathways in musculoskeletal neoplasia: involvement of the Indian Hedgehog-parathyroid hormone-related protein pathway. Pediatr Res 2003; 53: 539–543.
Rohatgi R, Milenkovic L, Corcoran RB, Scott MP . Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci USA 2009; 106: 3196–3201.
Barrett TL, Smith KJ, Hodge JJ, Butler R, Hall FW, Skelton HG . Immunohistochemical nuclear staining for p53, PCNA, and Ki-67 in different histologic variants of basal cell carcinoma. J Am Acad Dermatol 1997; 37: 430–437.
Kayaselçuk F, Zorludemir S, Gümürdülü D, Zeren H, Erman T . PCNA and Ki-67 in central nervous system tumors: correlation with the histological type and grade. J Neurooncol 2002; 57: 115–121.
Roy S, Ingham PW . Hedgehogs tryst with the cell cycle. J Cell Sci 2002; 115: 4393–4397.
Adams JM, Cory S . The Bcl-2 protein family: arbiters of cell survival. Science 1998; 281: 1322–1326.
Fedorko ME, Hirsch JG, Cohn ZA . Autophagic vacuoles produced in vitro I. Studies on cultured macrophages exposed to chloroquine. J Cell Biol 1968; 38: 377–391.
Ogata M, Hino S-i, Saito A, Morikawa K, Kondo S, Kanemoto S et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26: 9220–9231.
Bursch W . The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ 2001; 8: 569.
Jimenez-Sanchez M, Menzies FM, Chang Y-Y, Simecek N, Neufeld TP, Rubinsztein DC . The Hedgehog signalling pathway regulates autophagy. Nat Commun 2012; 3: 1200.
Wang Y, Han C, Lu L, Magliato S, Wu T . Hedgehog signaling pathway regulates autophagy in human hepatocellular carcinoma cells. Hepatology 2013; 58: 995–1010.
Milla LA, González-Ramírez CN, Palma V . Sonic Hedgehog in cancer stem cells: a novel link with autophagy. Biol Res 2012; 45: 223–230.
Mathew R, Karantza-Wadsworth V, White E . Role of autophagy in cancer. Nat Rev Cancer 2007; 7: 961–967.
Janku F, McConkey DJ, Hong DS, Kurzrock R . Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol 2011; 8: 528–539.
Zeng X, Kinsella TJ . Impact of autophagy on chemotherapy and radiotherapy mediated tumor cytotoxicity:’to live or not to live’. Front Oncol 2011; 1: 30.
Merimsky O, Bernstein-Molho R, Sagi-Eisenberg R . Targeting the mammalian target of rapamycin in myxoid chondrosarcoma. Anticancer Drugs 2008; 19: 1019–1021.
Wang Y, Ding Q, Yen C-J, Xia W, Izzo JG, Lang J-Y et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 2012; 21: 374–387.
El-Ashry D, Miller DL, Kharbanda S, Lippman ME, Kern FG . Constitutive Raf-1 kinase activity in breast cancer cells induces both estrogen-independent growth and apoptosis. Oncogene 1997; 15: 423.
Martin P, Poggi MC, Chambard JC, Boulukos KE, Pognonec P . Low dose cadmium poisoning results in sustained ERK phosphorylation and caspase activation. Biochem Biophys Res Commun 2006; 350: 803–807.
Sperandio S, Poksay K, De Belle I, Lafuente M, Liu B, Nasir J et al. Paraptosis: mediation by MAP kinases and inhibition by AIP-1/Alix. Cell Death Differ 2004; 11: 1066–1075.
Alvarez R, Elbashir S, Borland T, Toudjarska I, Hadwiger P, John M et al. RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob Agents Chemother 2009; 53: 3952–3962.
Tang G-Q, Yan T-Q, Guo W, Ren T-T, Peng C-L, Zhao H et al. (−)-Epigallocatechin-3-gallate induces apoptosis and suppresses proliferation by inhibiting the human Indian Hedgehog pathway in human chondrosarcoma cells. J Cancer Res Clin Oncol 2010; 136: 1179–1185.
Zhang N, Qi Y, Wadham C, Wang L, Warren A, Di W et al. FTY720 induces necrotic cell death and autophagy in ovarian cancer cells: A protective role of autophagy. Autophagy 2010; 6: 1157–1167.
Nopparat C, Porter JE, Ebadi M, Govitrapong P . The mechanism for the neuroprotective effect of melatonin against methamphetamine-induced autophagy. J Pineal Res 2010; 49: 382–389.
Kimura S, Fujita N, Noda T, Yoshimori T . Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol 2009; 452: 1–12.
Acknowledgements
We appreciate the Health Science Center of Peking University for providing experimental instruments and technical assistance. This work was supported by the Natural Science Foundation of China (No. 81072189, 81272381, and 81001193).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Piacentini
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sun, Y., Guo, W., Ren, T. et al. Gli1 inhibition suppressed cell growth and cell cycle progression and induced apoptosis as well as autophagy depending on ERK1/2 activity in human chondrosarcoma cells. Cell Death Dis 5, e979 (2014). https://doi.org/10.1038/cddis.2013.497
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.497
Keywords
This article is cited by
-
Structure–activity relationships for the G-quadruplex-targeting experimental drug QN-302 and two analogues probed with comparative transcriptome profiling and molecular modeling
Scientific Reports (2024)
-
GANT-61 induces cell cycle resting and autophagy by down-regulating RNAP III signal pathway and tRNA-Gly-CCC synthesis to combate chondrosarcoma
Cell Death & Disease (2023)
-
Constitutive GLI1 expression in chondrosarcoma is regulated by major vault protein via mTOR/S6K1 signaling cascade
Cell Death & Differentiation (2021)
-
GANT-61 Induces Autophagy and Apoptosis in Glioblastoma Cells despite their heterogeneity
Cellular and Molecular Neurobiology (2021)
-
MYC-regulated lncRNA NEAT1 promotes B cell proliferation and lymphomagenesis via the miR-34b-5p-GLI1 pathway in diffuse large B-cell lymphoma
Cancer Cell International (2020)