Abstract
Paired-like homeodomain 2 (Pitx2), first identified as the gene responsible for the Axenfeld–Rieger syndrome, encodes a protein factor that, controlling cell proliferation in a tissue-specific manner, has a crucial role in morphogenesis. During embryonic development, Pitx2 exerts a role in the expansion of muscle progenitors and is expressed at all stages of myogenic progression. In this study, we show that Pitx2 is phosphorylated by the protein kinase Akt2 and is necessary to ensure proper C2C12 myoblast proliferation and differentiation. Pitx2 associates with a ribonucleoprotein complex that includes the mRNA stabilizing factor HuR and sustains Ccnd1 (also known as Cyclin D1) expression, thereby prolonging its mRNA half-life. When the differentiation program is initiated, phosphorylation by Akt2 impairs the ability of Pitx2 to associate with the Ccnd1 mRNA-stabilizing complex that includes HuR and, as a consequence, Ccnd1 mRNA half-life is shortened. We propose that unphosphorylated Pitx2 is required to favor HuR-mediated Ccnd1 mRNA stabilization, thus sustaining myoblast proliferation. Upon Akt2-phosphorylation, the complex Pitx2/HuR/Ccnd1 mRNA dissociates and Ccnd1 mRNA is destabilized. These events contribute to the switch of C2C12 cells from a proliferating to a differentiating phenotype.
Similar content being viewed by others
Main
Paired-like homeodomain 2 (Pitx2) is a member of the bicoid class of paired homeodomain proteins that is mutated in patients affected by the Axenfeld–Rieger syndrome (ARS), an autosomal dominant haploinsufficient disorder that includes tooth anomalies, anterior segment eye defects, umbilical abnormalities, and facial dysmorphologies as cardinal features.1 The phenotype of mice with targeted deletion of Pitx2 mimics, in part, Axenfeld–Rieger syndrome ,2, 3, 4, 5 and its analysis established the critical role of Pitx2 in the development of cranio-facial structures, and of multiple organs including eye, pituitary, heart, and lung. Pitx2 is expressed in myotomes and migrating myoblasts3, 6, 7 and has been implicated in growth and survival of branchiomeric muscle progenitors.7, 8, 9 At early developmental stages, Pitx2 has been involved in the establishment of left–right asymmetry.10, 11, 12, 13
We previously showed that Pitx2 activates the transcription of specific growth-regulating genes such as those encoding Ccnd1 and Ccnd2 (also known as Cyclins D1 and D2) and it is also profoundly involved in the stabilization of the corresponding messengers during Wnt activation.6, 14 Moreover, Pitx2 mRNA is stabilized by Wnt activation, and this effect is mediated by AU-rich elements (AREs) in its 3′ untranslated region (3′UTR).14 We showed that Pitx2 protein is required for stabilization of its own mRNA, as well as of those encoding Ccnd1 and Ccnd2.14 This effect is achieved, in part, by modulating the function of the ARE-binding protein (ARE-BP) HuR as Pitx2 and HuR belong to the same complex.14
It is now clear that mRNA decay regulation by different signals makes a huge contribution to the global control of gene expression.15 AREs, located in the 3′UTR of many short-lived transcripts, promote mRNA deadenylation, decapping, and degradation, with ARE-BPs functioning as trans-acting factors responsible for the modulation of decay rates.16, 17 Some ARE-BPs are decay-promoting factors (TTP, BRF1, KSRP) and others, such as HuR, are stabilizing factors.16
Skeletal muscle formation or myogenesis is a complex and highly regulated process that involves the proliferation of myoblasts, followed by morphological, biochemical, and molecular modifications, which result in the formation of multinucleated myotubes.18, 19, 20 The C2C12 myogenic cell line, derived from regenerating adult mouse skeletal muscle, provides a good in vitro model for studying the major steps of myoblast proliferation and differentiation (21 and literature cited therein). We have reported that the tight control of the decay rates of transcripts encoding several pro-myogenic factors, in addition to their transcriptional regulation, is essential to ensure that the correct differentiation program takes place.21, 22, 23 HuR, regulating the expression of the mRNAs encoding the pro-myogenic transcription factors MyoD and myogenin has a key role in muscle fiber formation.21, 22 As HuR and Pitx2 associate to stabilize target transcripts,14 we hypothesized that the two proteins could cooperate to affect the fate of myoblasts during the myogenic process.
In this study, we report that Pitx2 is necessary to ensure proper C2C12 myoblast proliferation and differentiation. Pitx2 and HuR belong to the same ribonucleoprotein complex that controls the decay rate of the Ccnd1 mRNA, and this function is regulated by Pitx2 phosphorylation by Akt2.
Results
Pitx2 is phosphorylated by Akt2 in C2C12 myoblasts
In a screening for protein serine/threonine kinases able to phosphorylate in vitro Pitx2 (Gherzi R, unpublished results), we found that Akt2 is able to phosphorylate, in a concentration-dependent manner, the purified recombinant Pitx2c portion encompassing amino acids 1–160, whereas it does not phosphorylate the C-terminal part of the protein including amino acids 161–324 (Figure 1a, left panel). An in silico search (http://scansite.mit.edu/) for potential Akt2 phosphorylation sites indicated threonine 97, embedded in a nearly perfect consensus sequence (RQRTHFT), as the most probable Akt2 target residue. Indeed, Akt2 failed to phosphorylate the mutant Pitx2c 1–160 in which threonine 97 was substituted with alanine (Pitx2c 1–160T97A, Figure 1a, right panel). The sequence RQRTHFT is located in the homeodomain, is present in the three Pitx2 splicing variants, and is evolutionarily conserved from Drosophila to human (Figure 1b). As expected, Akt2 is able to phosphorylate in vitro Pitx2a and b (data not shown). Akt1 does not phosphorylate Pitx2c in vitro, although it phosphorylates KSRP, which is used as a control (24 Figure 1c). Interestingly, human PITX2A is phosphorylated by PKC in 10 different residues including threonine 44, which corresponds to mouse Pitx2c threonine 97.25
Pitx2 is phosphorylated in Threonine 97 by Akt2. (a) Purified recombinant GST-Pitx2c 1–160 (lanes 1–5, 8), purified recombinant GST-Pitx2c 161–324 (lane 6), or purified recombinant GST-Pitx2c 1–160 T97A (lane 9) were incubated with either active recombinant Akt2 in kinase buffer (lanes 1–4 and 6–9) or buffer alone (lane 5) in the presence of γ[32P]ATP. Labeled proteins were separated by SDS-PAGE, blotted to nylon membranes, and detected by autoradiography. Open arrows point to the position of phosphorylated recombinant Akt2, black arrows point to the position of Pitx2c 1–160 and Pitx2c 161–320. (b) Sequence comparisons of the potential AKT phosphorylation site in the three murine Pitx2 splice variants and in Pitx genes from different species (mmu, Mus musculus; hsa, Homo sapiens; gga, Gallus gallus; dre, Danio rerio; dme, Drosophila melanogaster). The symbol of the phosphorylated threonine is in bold. (c) Purified recombinant GST-Pitx2c 1–160 (lanes 1–4, 9), purified recombinant GST-Pitx2c 161–324 (lane 6), or purified recombinant KSRP23, 24 (lanes 7, 8) were incubated with either active recombinant Akt1 in kinase buffer (lanes 1–7), buffer alone (lane 8), or active recombinant Akt2 in kinase buffer (lane 9) in the presence of γ[32P]ATP. Labeled proteins were separated by SDS-PAGE, blotted to nylon membranes, and detected by autoradiography (upper panel). Open arrow points to the position of recombinant KSRP, black arrow points to the position of Pitx2c 1–160. The same membranes were subjected to immunoblotting using anti-GST antibody (lower panel). (d) In vivo [32P]orthophosphate metabolic labeling of C2C12 cells cultured either in GM (lanes 1, 3) or in DM (lanes 2, 4). Aliquots of the lysates were immunoprecipitated with protein A-/protein G-Sepharose bound with either preimmune serum (P.I, lanes 3, 4) or anti-Pitx2 guinea pig antibody (lanes 1, 2), separated by SDS-PAGE, and autoradiographed (upper panel). The same membranes were subjected to immunoblot analysis using purified anti-Pitx2 rabbit antibody (lower panel)
Considering the role of Pitx2 during muscle development8, 9 and the function of PI3K–Akt signaling in myoblast differentiation,26, 27 we chose to study the functional relevance of Pitx2 phosphorylation by Akt2 in C2C12 cells that differentiate into myotubes upon shift from growth medium (GM) to low-serum-containing differentiation medium (DM).23 As shown in Figure 1d, endogenous Pitx2 was phosphorylated in response to DM in intact C2C12 cells. Furthermore, constitutively active Akt2 induced 32P incorporation into Flag-tagged Pitx2c when coexpressed in proliferating C2C12 cells (Supplementary Figure S1a). Finally, Akt2 immunopurified from C2C12 cells, either maintained in GM or exposed to DM for different intervals of time, was used to phosphorylate Pitx2c 1–160 in vitro. As shown in Supplementary Figure S1b, the kinase activity of Akt2 toward Pitx2c was rapidly (1 h) enhanced by exposure to DM. Akt2 activity was maintained up to 36 h in DM and started declining between 36 and 48 h of differentiation, whereas the expression of the enzyme remained unchanged (Supplementary Figure S1b).
Altogether, these data show that Pitx2 is a substrate for Akt2 at early phases of C2C12 cell differentiation.
Pitx2 is required for C2C12 myoblast differentiation
To gain an insight into Pitx2 function in the course of C2C12 cell differentiation, we transiently knocked down the three Pitx2 isoforms by RNA interference (siRNA, Figure 2a). The expression of some myogenic differentiation markers, including Myog (also called myogenin), Ckm (also called muscle creatine kinase), Myod1, and Cdkn1a (also called p21), was severely impaired in siPitx2-transfected C2C12 cells induced to differentiate by DM (Figure 2b). On the contrary, the expression of an early marker of myoblast progenitors and satellite cells (Pax7), as well as markers of myogenic determination such as Myf5, and Myf6 (also called Mrf4)20 remained unchanged (Supplementary Figure S1c). Pax3, another marker of myoblast progenitors,20 is not expressed in C2C12 cells.
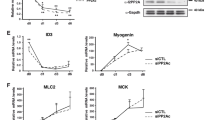
Pitx2 is required for C2C12 myoblast differentiation into myotubes. (a) Immunoblot analysis of total cell extracts from either siLuciferase (siCtrl) or siPitx2-transfected C2C12 cells performed to detect the indicated proteins. (b) Quantitative RT-PCR (qPCR) analysis of Myog, Ckm, Myod1, Cdkn1a transcripts in C2C12 cells transfected with either siLuciferase (siCtrl) or siPitx2 and cultured either in growth medium (GM) or differentiation medium (DM) for 44 h. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate. (c) Immunoblot analysis of total cell extracts from stable C2C12 clones transfected with either pSuper-Puro empty vector (mock-C2C12) or shPitx2 cloned into pSuper-Puro (C2C12-shPitx2). Cells were cultured in either growth medium (GM) or differentiation medium (DM) for 3 days. Antibodies are indicated below. Statistical significance: one asterisk P<0.01 (Student's t-test)
To extend our observations, we prepared a short-hairpin (sh) RNA-based vector targeting mouse Pitx2 and generated two independent stable C2C12 clones, in which approximately 70% of Pitx2 was knocked down (data not shown). The two clones yielded superimposable results and henceforth are collectively cited as C2C12-shPitx2 cells. We confirmed in C2C12-shPitx2 cells that Pitx2 is required for the proper expression of myogenic factors (Figure 2c, Supplementary Figure S1d, e), including members of the recently described class of myogenic miRNAs28 (Supplementary Figure S1f). Accordingly, connexin 43, a described target of miR-206, was upregulated in C2C12-shPitx2 cells (28 and literature cited therein; Supplementary Figure S1g).
Our data indicate that Pitx2 is required for differentiation of C2C12 myoblasts into myotubes.
Pitx2 controls C2C12 myoblast proliferation and affects Ccnd1 mRNA decay
We previously reported that Pitx2 controls cell proliferation in specific tissues.6 Here, we found that Pitx2 knockdown causes a growth reduction in C2C12 cells cultured in GM as assessed by crystal violet staining (Supplementary Figure S2a). Using C2C12-shPitx2 cells, we confirmed by both BrdU incorporation into DNA and crystal violet staining that Pitx2 knockdown reduces the proliferation of exponentially growing myoblasts, whereas it does not affect the proliferation rate of cells cultured in DM (Figure 3a and b). Conversely, Pitx2 knockdown did not induce apoptosis in C2C12 cells (Supplementary Figure S2b).
Pitx2 modulates C2C12 myoblast proliferation and controls the Ccnd1 mRNA decay rate. (a, b) BrdU staining (a) or crystal violet staining (b) of stable C2C12 clones transfected with either pSuper-Puro empty vector (mock C2C12) or shPitx2 cloned into pSuper-Puro (C2C12-shPitx2). Cells were cultured in either growth medium (GM) or differentiation medium (DM) for 3 days. The values shown in (b) are averages (±S.E.M.) of three independent experiments performed in triplicate. (c) Immunoblot analysis of total cell extracts from either siLuciferase (siCtrl) or siPitx2-transfected C2C12 cells performed to detect the indicated proteins. (d) Immunoblot analysis of either nuclear (nuc) or cytoplasmic (cyto) extracts from C2C12 cells cultured in either growth medium (GM) or differentiation medium (DM) for 3 days. The detected proteins are indicated on the right. (e) Quantitative RT-PCR (qPCR) analysis of Ccnd1 and Myog transcripts in C2C12 cells transfected with either siLuciferase (siCtrl) or siPitx2 and cultured in growth medium (GM). Total RNA was isolated at the indicated times after the addition of DRB. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate. (f) C2C12 cells were cultured in either growth medium (GM) or differentiation medium (DM) for the indicated intervals of time, lysed, and total cell extracts were immunoprecipitated as indicated. RNA was purified from immunocomplexes and analyzed by qPCR to detect Ccnd1 mRNA. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate. Statistical significance: one asterisk P<0.01 (Student's t-test)
In agreement with our previous findings,6, 14 Pitx2 knockdown reduced the expression of Ccnd1 in proliferating C2C12 myoblasts (Figure 3c, Supplementary Figure S2c), whereas it did not affect Ccnd1 expression in differentiating C2C12 cells (Supplementary Figure S2d).
Surprisingly, we found that Pitx2 is predominantly cytoplasmic in C2C12 cells and this observation was confirmed by using two different anti-Pitx2 antibodies (Figure 3d and data not shown). We previously reported that Pitx2 is able to controls the expression of Ccnd1 (and other transcripts) at both transcriptional and posttranscriptional levels.6, 14 The subcellular localization of Pitx2 in C2C12 cells prompted us to investigate the consequences of Pitx2 knockdown on the decay rate of Ccnd1 mRNA in these cells. In vitro degradation assays revealed that Ccnd1 mRNA is stable in proliferating C2C12 myoblasts and its half-life is reduced by serum withdrawal-induced differentiation (Supplementary Figure S3a). These data are consistent with the reduction in Ccnd1 mRNA levels that we observed in the course of C2C12 cell differentiation (Supplementary Figure S3b). Most importantly, transient Pitx2 knockdown significantly shortened the Ccnd1 transcript half-life in intact proliferating C2C12 myoblasts, whereas the decay rate of Myog mRNA was unaffected (Figure 3e). Similar results were obtained on stable Pitx2 knockdown using in vitro degradation assays (Supplementary Figure S3c).
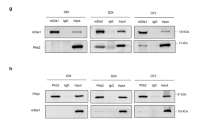
We have previously shown that, although it is not able to directly bind to mRNA, Pitx2 participates in a ribonucleoprotein complex containing, among others, the mRNA stabilizing factor HuR (14 Briata P, unpublished). Furthermore, HuR has been reported to target the inherently labile Ccnd1 transcript and to induce its stabilization.29, 30 By ribonucleoprotein complex immunoprecipitation (RIP) analysis, we found that anti-Pitx2 antibody immunoprecipitates a Ccnd1 mRNA-containing ribonucleoprotein complex from C2C12 cell extracts and that this interaction is reduced by cell differentiation (Figure 3f upper panel). Similarly, HuR binding to Ccnd1 mRNA is reduced in differentiated C2C12 myotubes when compared with proliferating C2C12 myoblasts (Figure 3f lower panel).
These results suggest that Pitx2 modulates cell proliferation and Ccnd1 mRNA half-life by participating in a ribonucleoprotein complex that contains HuR.
Phosphorylation by Akt2 impairs the ability of Pitx2 to associate with a Ccnd1 mRNA-stabilizing complex
On the basis of the above results, we investigated whether HuR–Ccnd1 mRNA interaction depends on Pitx2. As shown in Figure 4a, transient Pitx2 knockdown reduced the interaction of HuR with Ccnd1 mRNA in proliferating C2C12 cells (upper panel) without affecting HuR expression levels (lower panel).
Dynamic participation of Pitx2 in a ribonucleoprotein complex stabilizing Ccnd1 mRNA. (a) C2C12 cells were transfected with either siLuciferase (siCtrl) or siPitx2, cultured in growth medium, and lysed. Total cell extracts were immunoprecipitated as indicated. RNA was purified from immunocomplexes and analyzed by qPCR to detect Ccnd1 mRNA. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate (upper panel). Immunoblot analysis of HuR in total extracts of C2C12 cells transfected with either siLuciferase (siCtrl) or siPitx2 and cultured in growth medium (lower panel). (b) qPCR analysis of Myog and Ccnd1 transcripts in C2C12 cells infected with an adenoviral vector expressing either a control empty adenoviral vector (Ad-Control) or an adenoviral vector expressing constitutively active myristilated AKT2 (Ad-myrAKT2) cultured in either growth medium (GM) or differentiation medium (DM) for 48 h. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate. (c) C2C12 cells were infected with an adenoviral vector expressing either a control empty adenoviral vector (Ad-Control) or an adenoviral vector expressing constitutively active myristilated AKT2 (Ad-myrAKT2) and cultured in growth medium. Cells were lysed, total cell extracts were immunoprecipitated as indicated, RNA was purified from immunocomplexes, and analyzed by qPCR to detect Ccnd1 mRNA. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate. (d) GST pull-down of endogenous 14-3-3 and HuR from C2C12 cell total extracts using either control GST, GST-Pitx2c 1–160 (GST-Pitx2c), in vitro phosphorylated GST-Pitx2c 1–160 (GST-pPitx2c), or in vitro phosphorylated GST-Pitx2c 1–160T97A (GST-pPitx2cT97A). Proteins were analyzed by immunoblotting using either anti-14-3-3 or anti-HuR antibodies. (e, f) Co-immunoprecipitation of Pitx2 and either HuR (e) or 14-3-3 in C2C12 cells cultured in either GM or DM. Cell lysates were immunoprecipitated as indicated and analyzed by immunoblotting using the indicated antibodies. P.I. indicates preimmune serum. Statistical significance: one asterisk P<0.01 (Student's t-test)
PI3K–Akt activation favors the differentiation of myoblasts into myotubes (26 and Figure 4b left panel). Importantly, forced activation of Akt2 induced a significant reduction in Ccnd1 mRNA levels in C2C12 cells, reducing the half-life of Ccnd1 mRNA (Figure 4b right panel, Supplementary Figure S3d). Furthermore, Akt2 activation decreased the ability of Pitx2 to participate in the Ccnd1 stabilizing complex (Figure 4c, upper panel) and of HuR to interact with Ccnd1 mRNA (Figure 4c, lower panel).
It is well known that phosphorylation by Akt kinases can change the ability of substrate proteins to interact with molecular partners.31 By GST pull-down experiments, we found that phosphorylation by Akt2 increases the ability of Pitx2c to interact with the multifunctional protein 14-3-3 while decreasing its ability to interact with HuR (Figure 4d). Under the same conditions, Pitx2cT97A, which does not undergo Akt2 phosphorylation (Figure 1a), was unable to interact with 14-3-3, whereas it associated with HuR (Figure 4d). Accordingly, coimmunoprecipitation experiments revealed that Pitx2 interaction with HuR was strongly reduced in C2C12 cells cultured in DM, whereas Pitx2 interaction with 14-3-3 was increased (Figure 4e and f, and Supplementary Figure S3e).
Next, we investigated whether the expression of Pitx2cT97A in C2C12 cells (Supplementary Figure 3f) interferes with their differentiation. First, RIP experiments revealed that Ccnd1 mRNA remained associated with Pitx2cT97A expressed in C2C12 cells on differentiation induction (Figure 5a). In keeping with these observations, Pitx2cT97A remained associated with HuR on shift of C2C12 cells to DM (Figure 5b). As shown in Figure 5c, DM failed to downregulate Ccnd1 mRNA and induce myogenin mRNAs in C2C12 cells expressing Pitx2cT97A.
T97A Pitx2c mutant is maintained in a ribonucleoprotein complex including Ccnd1 mRNA and HuR upon shifting C2C12 cells culture medium to DM. (a) C2C12 cells were transfected with either Flag-Pitx2c or Flag-Pitx2cT97A, cultured in either growth medium (GM) or differentiation medium (DM) for 24 h, lysed, and total cell extracts were immunoprecipitated as indicated. RNA was purified from immunocomplexes and analyzed by qPCR to detect Ccnd1 mRNA. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate. (b) Co-immunoprecipitation of transfected Flag-Pitx2c or Flag-Pitx2cT97A and HuR in C2C12 cells cultured in either GM or DM for 24 h. Cell lysates were immunoprecipitated as indicated and analyzed by immunoblotting using anti-HuR. cIgGs are control mouse IgGs. (c) Quantitative RT-PCR (qPCR) analysis of Myog and Ccnd1 transcripts in C2C12 cells transfected with either pCMV-TAG2B empty vector (mock) or Flag-Pitx2cT97A and cultured either in growth medium (GM) or differentiation medium (DM) for 24 h. The values shown are averages (±S.E.M.) of three independent experiments performed in triplicate. (d) A model for control of Ccnd1 mRNA decay by Akt2-dependent Pitx2 phosphorylation in C2C12. The asterisk marks a decay-promoting complex. Statistical significance: one asterisk P<0.01 (Student's t-test)
Altogether, our data indicate that Pitx2 is a required factor for HuR–Ccnd1 mRNA interaction in proliferating C2C12 myoblasts and that Akt2 activation, inducing Pitx2 phosphorylation, causes its dismissal from the Ccnd1 mRNA-stabilizing complex.
Discussion
In this report, we show that Pitx2 is necessary to ensure proper proliferation of C2C12 myoblasts under high-serum culture conditions, as well as for their differentiation under low-serum culture conditions. In C2C12 cells, Pitx2 predominantly localizes in the cytoplasm and is part of an mRNA-stabilizing complex, including the ARE-BP HuR, which controls the Ccnd1 mRNA half-life. On Pitx2 phosphorylation by Akt2, the Pitx2/HuR-containing complex is dismissed from Ccnd1 mRNA leading to destabilization of the transcript.
The contribution of different Akt isoforms to myoblast differentiation is still debated.32 We found that Akt1 does not phosphorylate Pitx2c in vitro. This result is consistent with previous observations of isoform-specific phosphorylation of cellular substrates by Akt family members33, 34 and with the reported preference for an arginine residue at position-5 (absent in Pitx2c Akt recognition consensus site) as displayed by Akt1.35
It has been shown that Pitx2 is implicated in myogenesis during mouse embryonic development.8, 9 In this study, we propose that, in proliferating C2C12 myoblasts, Pitx2 is required to maintain an adequate rate of cell division. A similar role has been suggested for Pax3 and Pax7 in satellite cell-derived myoblasts.36 Pax3 is not expressed in C2C12 cells, whereas Pax7 expression is not affected by Pitx2 knockdown, thus suggesting that Pitx2 and Pax7 may regulate parallel pathways that converge on common target genes. However, it is noteworthy that Pitx2 knockdown is sufficient to severely impair the expression of myogenic differentiation markers such as Myog, Myod1, Ckm, Cdkn1a, as well as myogenic miRNAs. We propose that Pitx2, in concert with Pax3 and Pax7, keeps myoblasts actively proliferating and poised for differentiation in response to appropriate environmental conditions. In our hypothesis, Pitx2 promotes C2C12 cell expansion, while maintaining commitment to the myogenic lineage.
We previously reported that Pitx2 can exert a dual role regulating the expression of target genes not only transcriptionally but also posttranscriptionally, being able to modulate the turnover rate of select ARE-containing mRNAs, including Ccnd1, in response to Wnt/β-catenin activation.14 We now report that, in C2C12 cells, Pitx2 displays a predominant cytoplasmic localization and this observation prompted us to analyze in detail the role of Pitx2 on Ccnd1 mRNA turnover. Ccnd1 mRNA is rather stable in proliferating C2C12 myoblasts, whereas it is destabilized on serum withdrawal-induced differentiation. Pitx2 seems to participate in these events because its knockdown reduces Ccnd1 transcript half-life and steady-state levels in proliferating C2C12 myoblasts. Studies conducted in Gorospe's laboratory reported that Ccnd1 mRNA is a direct target of the stabilizing ARE-BP HuR.30 Indeed, we observed that HuR interaction with Ccnd1 mRNA is modulated by differentiation. We have previously suggested a role for Pitx2 in controlling HuR function in the context of a stabilizing ribonucleoprotein complex.14 Our present evidence, suggesting that Pitx2 knockdown impairs the ability of HuR to interact with Ccnd1 mRNA, emphasizes the role of Pitx2 as a required component of the HuR-containing stabilizing ribonucleoprotein complex.
It has been shown that activation of the PI3K-Akt pathway strongly stimulates myogenesis, greatly increasing myotube formation in myoblast cultures and upregulating muscle-specific proteins.26, 27 Here, we propose that Akt2-induced downregulation of Ccnd1 expression through destabilization of its mRNA represents an additional mechanism by which the signaling pathway controls the transition from the undifferentiated to the differentiated state in C2C12 cells. We previously reported that the PI3K-Akt pathway can affect the decay rate of ARE-containing transcripts regulating the function of relevant ARE-BPs.24 Once phosphorylated by Akt1/2, KSRP and Brf1, two destabilizing ARE-BPs, interact with the multifunctional protein 14-3-3 and, in turn, their function is compromised.24, 37 HuR is not a substrate for Akt kinases (Gherzi R, unpublished observation) and we show that Akt2 activation modulates the function of HuR indirectly, through Pitx2 phosphorylation.
The T97A Pitx2c mutant, which is not phosphorylated by Akt2, is present in a ribonucleoprotein complex including HuR and Ccnd1 mRNA on shifting C2C12 culture medium to DM and, as a consequence, Ccnd1 expression is not downregulated. Interestingly, myogenin mRNA levels are not induced in the same cells. These results strengthen the idea that modulation of Ccnd1 mRNA levels by Pitx2c is one of the mechanisms underlying C2C12 myoblast differentiation. Indeed, reduction of Ccnd1 transcript on specific knockdown in proliferating C2C12 myoblasts reduced the expression of some muscle-specific transcripts (Briata P, unpublished), thus underscoring the requirement of a proper proliferation rate for the subsequent full differentiation.
We propose that, in proliferating C2C12 myoblasts, Pitx2 interacts with HuR favoring its binding to Ccnd1 mRNA, which becomes more stable. When cells are shifted to DM, Akt2-phosphorylated Pitx2 fails to interact with HuR, whereas it associates with 14-3-3. Consequently, HuR no longer binds to Ccnd1 mRNA, whose half-life is shortened. This mechanism (summarized in the model presented in Figure 5d) contributes to maintain a high proliferation rate in C2C12 cells cultured in GM and to arrest cell proliferation on shift to DM.
Materials and Methods
Cell cultures, transfections, plasmids, and adenoviral infections
Murine C2C12 myoblasts were cultured in DMEM plus 20% FBS (Growth Medium, GM). Myogenic differentiation was induced by incubation in DMEM plus 2% horse serum (Differentiation Medium, DM). For transient transfections, C2C12 cells were electroporated using Nucleofector II, according to the manufacturer's instructions (Amaxa, Walkersville, MD, USA). C2C12 cell transfections that were aimed at obtaining stable shPitx2 transfectants were performed using Lipofectamine Plus (Invitrogen, Carlsbad, CA, USA), and Puromycin (Invivogen, San Diego, CA, USA) was used at 2.5 μg/ml for selection. Clones of transfected cells were selected by limiting dilution and were used for experiments. Full-length mouse Pitx2c cDNA was cloned in pCMV–TAG2B (Stratagene, La Jolla, CA, USA) obtaining Flag-Pitx2c. To obtain Flag-Pitx2cT97A, we mutagenized Flag-Pitx2c using a QuikChange II site-directed mutagenesis kit (Stratagene) and the following primers 5′-CCGGCAGAGGGCTCATTTCACTAGCCAG-3′, 5′-CTGGCTAGTGAAATGAGCCCTCTGCCGG-3′. Stable myrAKT-C2C12 transfectants have been described previously.24 Adenoviral vectors pAdPitx2c and pAdmyrAKT2 were purchased from Vector Biolabs (Eagleville, PA, USA) and were used as previously described.23
Recombinant proteins and antibodies
Production and purification of recombinant GST-Pitx2c 1–160, GST-Pitx2c 161–324, and GST-Pitx2c 1–160T97A were performed as previously described.14 Affinity-purified rabbit polyclonal anti-Pitx2 antibody was purchased from Capra Science (Angelholm, Sweden). Anti-Pitx2 polyclonal antibody raised in guinea-pig has been described previously.6 Mouse monoclonal anti-HuR 3A2 antibody has been described previously.38 Mouse monoclonal anti-myogenin (F5D) and anti-MHC antibodies (MF20) were from Developmental Studies Hybridoma Bank University of Iowa. Mouse monoclonal antibody anti-α-tubulin and rabbit polyclonal antibodies anti-connexin 43 and anti-HDAC1 were from Sigma (St. Louis, MO, USA). Affinity-purified mouse monoclonal anti-Cyclin D1/2 (cat no. 05-362) and rabbit polyclonal anti-GST antibody were from Millipore (Billerica, MA, USA). Anti-AKT rabbit polyclonal antibody was from Cell Signaling. Affinity-purified rabbit polyclonal anti–14-3-3β (K-19, which recognizes all 14-3-3 family members) was from Santa Cruz (Santa Cruz, CA, USA).
siRNA and shRNA-mediated Pitx2 knockdown
To knock down mouse Pitx2, 5′-UGGAGAAAGCGGGAACGCA-3′ siRNA was used and to knock down mouse Ccnd1, 5′-GUUGUGCAUCUACACUGAC-3′ siRNA was used; control Luciferase siRNA was 5′-CGUACGCGGAAUACUUCGAUU -3′ (synthesized by TIB MolBiol, Genova, Italy). To stably knock down mouse Pitx2, the following oligonucleotide 5′-UGGAGAAAGCGGGAACGCA-3′ was cloned into pSUPER-Puro (Oligoengine, Seattle, WA, USA), following the manufacturer's instructions.
In vitro kinase assays and 32P orthophosphate metabolic labeling
AKT1 and AKT2 kinase assays were performed using preactivated enzymes purchased from Millipore (Billerica, MA, USA) as recommended by the manufacturer. [γ-32P]ATP (3000 Ci/mmol) was from Perkin Elmer. In vivo 32P orthophosphate metabolic labeling of C2C12 cells was performed as previously23, 24 described, incubating cells with orthophosphate for 16 h.
Quantitative RT-PCR
Total RNA was isolated using the miRNeasy mini kit (Qiagen, Milano, Italy), treated with DNAseI (Promega, Madison, WI, USA), and retro-transcribed using SuperScript III (Invitrogen) according to the manufacturer's instructions. For quantitative RT-PCR (qPCR), 100 ng of DNAse I-treated total RNA was retro-transcribed and PCR reactions were performed using Real Mastermix (5 Prime, Hamburg, Germany) and the Realplex II Mastercycler (Eppendorf, Milano, Italy). The sequence-specific primers used for PCR reactions are listed in Supplementary Table S1. Quantitative PCR analysis of mature miRNAs (qmiR-PCR) was performed using the NCode miRNA first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions and the IQ Sybr Green Mix Super (Bio-Rad, Hercules, CA, USA).
Northern blot analysis
Total RNA (10 μg/lane) was resolved on 15% polyacrylamide-urea gels, and electroblotted onto HyBond N+ membranes (GE Healthcare, Buckinghamshire, England). Membranes were hybridized overnight with radiolabeled antisense miRNAs in ExpressHyb solution (Clontech, Mountain View, CA, USA). After hybridization, membranes were washed three times with 2X SSC and 0.05% SDS, twice with 0.1 × SSC and 0.1% SDS, exposed overnight to imaging screens, and analyzed using a Storm 860 PhosphorImager (GE Healthcare). Signals were quantitated using Imagequant V1.2 (GE Healthcare). The same blot was hybridized (on stripping in boiling 0.1% SDS) with three distinct probes, including control U6 RNA.
RNA in vitro degradation and measure of mRNA half-life in intact cells
32P-labeled RNAs were synthesized and used as substrates for in vitro degradation assays as reported.39 ARECcnd1 and the E3 sequence from Pitx2 3′UTR have been described previously.14 Cells under different culture conditions were treated with 100 μM DRB (5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole, 5,6-Dichlorobenzimidazole riboside), harvested at the indicated times, and total RNA was isolated and analyzed as above.
Ribonucleoprotein complexes immunoprecipitation (RIP)
RIP was performed as previously described by Chen et al.39 with minor modifications.40 Briefly, cell lysates were immunoprecipitated with either Protein A- or ProteinA/ProteinG-Sepharose-coupled antibodies at 4°C overnight. Pellets were sequentially washed with the following buffers: buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl); buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl); and buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1). Total RNA was prepared using Trizol (Invitrogen), retro-transcribed using random primers, and amplified by qPCR. The primer sequences are detailed in Supplementary Table SI.
Proliferation and apoptosis assays
5-Bromo-2′-deoxy-uridine labeling was performed in C2C12 cells using a commercial kit and following the manufacturer's instructions (Roche, Penzberg, Germany). Crystal violet staining was performed on formalin-fixed C2C12 cells for 30 min at room temperature. The dye was solubilized in 10% acetic acid and absorbance was measured at 590 wavelength. Cleaved caspase-3 immunoblot detection was performed using a commercial kit and following the manufacturer's instructions (Cell Signaling, Danvers, MA, USA).
Nuclear and cytoplasmic fractionation, S100 extracts preparation
Nuclear and cytoplasmic cell extracts were performed using the NE-PER commercial kit and following the manufacturer's instructions (Pierce, Rockford, IL, USA). S100 extracts were prepared as previously described.39
GST pull-down assays
Escherichia coli, transformed with pGEX-based recombinant plasmids (GE Healthcare), were grown until A600 reached 0.6, were induced with 1 mM of isopropyl-β-D-thiogalactoside (IPTG) for 3 h, harvested, washed with PBS, and lysed by sonication. After centrifugation (14 000 × g for 15 min at 4°C), the supernatants were incubated with glutathione-Sepharose beads (GE Healthcare) for 1 h at 4°C under rotation. Beads were washed four times with pull-down buffer (PDB 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 0.2% Nonidet P-40, 10% glycerol, and a mixture of protein inhibitors (Complete, Roche)), saturated with bovine serum albumin, washed again with PDB, and incubated with 800 μg of cell extracts (2 h, 4°C, rotating). Beads, collected by centrifugation (30 s 5000 × g), were washed 4 times with PDB. Bound proteins were eluted in SDS sample buffer, resolved by SDS-PAGE, and analyzed by immunoblot.
Abbreviations
- ARS:
-
Axenfeld-Rieger syndrome
- ARE:
-
AU-rich element
- ARE-BP:
-
ARE-binding protein
- 3′UTR:
-
3′ untranslated region
- GM:
-
growth medium
- DM:
-
differentiation medium
- BrdU:
-
5-Bromo-2′-deoxy-uridine
- RIP:
-
ribonucleoprotein complexes immunoprecipitation
- DRB:
-
5,6-Dichloro-1-b-D-ribofuranosylbenzimidazole, 5,6-Dichlorobenzimidazole riboside)
References
Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet 1996; 14: 392–399.
Gage PJ, Suh H, Camper SA . Dosage requirement of Pitx2 for development of multiple organs. Development 1999; 126: 4643–4651.
Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 1999; 126: 5749–5758.
Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F et al. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 1999; 401: 279–282.
Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF . Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 1999; 401: 276–278.
Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T et al. Identification of a Wnt/Dvl/beta-Catenin → Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 2002; 111: 673–685.
L’Honoré A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P et al. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol 2007; 307: 421–433.
Dong F, Sun X, Liu W, Ai D, Klysik E, Lu MF et al. Pitx2 promotes development of splanchnic mesoderm-derived branchiomeric muscle. Development 2006; 133: 4891–4899.
Shih HP, Gross MK, Kioussi C . Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc Natl Acad Sci USA 2007; 104: 5907–5912.
Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y et al. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell 1998; 94: 299–305.
Logan M, Pagán-Westphal SM, Smith DM, Paganessi L, Tabin CJ . The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell 1998; 94: 307–317.
Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA . Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell 1998; 94: 319–324.
Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature 1998; 394: 545–551.
Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY et al. The Wnt/beta-catenin → Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell 2003; 12: 1201–1211.
Hao S, Baltimore D . The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 2009; 10: 281–288.
Wilusz CJ, Wilusz J . Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet 2004; 20: 491–497.
Houseley J, Tollervey D . The many pathways of RNA degradation. Cell 2009; 136: 763–776.
Yun K, Wold B . Skeletal muscle determination and differentiation: story of a core regulatorynetwork and its context. Curr Opin Cell Biol 1996; 8: 877–889.
Chargé SB, Rudnicki MA . Cellular and molecular regulation of muscle regeneration. Physiol Rev 2004; 84: 209–238.
Buckingham M . Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev 2006; 16: 525–532.
van der Giessen K, Di-Marco S, Clair E, Gallouzi IE . RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem 2003; 278: 47119–47128.
Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Muñoz-Canoves P et al. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol 2003; 23: 4991–5004.
Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M et al. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol Cell 2005; 20: 891–903.
Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C et al. 2006 The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol 2006; 5: e5.
Espinoza HM, Ganga M, Vadlamudi U, Martin DM, Brooks BP, Semina EV et al. Protein kinase C phosphorylation modulates N- and C-terminal regulatory activities of the PITX2 homeodomain protein. Biochemistry 2005; 44: 3942–3954.
Jiang BH, Zheng JZ, Vogt PK . An essential role of phosphatidylinositol 3-kinase in myogenic differentiation. Proc Natl Acad Sci USA 1998; 95: 14179–14183.
Xu Q, Wu Z . The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J Biol Chem 2000; 275: 36750–36757.
van Rooij E, Liu N, Olson EN . MicroRNAs flex their muscles. Trends Genet 2008; 24: 159–166.
Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M . Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 2001; 21: 5889–5898.
Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M . Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 2004; 23: 3092–3102.
Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 1997; 91: 961–971.
Héron-Milhavet L, Mamaeva D, Rochat A, Lamb NJ, Fernandez A . Akt2 is implicated in skeletal muscle differentiation and specifically binds Prohibitin2/REA. J Cell Physiol 2008; 214: 158–165.
Yamada E, Okada S, Saito T, Ohshima K, Sato M, Tsuchiya T et al. Akt2 phosphorylates Synip to regulate docking and fusion of GLUT4-containing vesicles. J Cell Biol 2005; 168: 921–928.
Kato S, Ding J, Du K . Differential activation of CREB by Akt1 and Akt2. Biochem Biophys Res Commun 2007; 354: 1061–1066.
Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P . Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett 1996; 399: 333–338.
Collins CA, Gnocchi VF, White RB, Boldrin L, Perez-Ruiz A, Relaix F et al. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS ONE 2009; 4: e4475.
Schmidlin M, Lu M, Leuenberger SA, Stoecklin G, Mallaun M, Gross B et al. The ARE-dependent mRNA destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J 2004; 23: 4760–4769.
Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA 2000; 97: 3073–3088.
Chen CY, Gherzi R, Andersen JS, Gaietta G, Jürchott K, Royer HD et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev 2000; 14: 1236–1248.
Ruggiero T, Trabucchi M, De Santa F, Zupo S, Harfe BD, McManus MT et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J 2009; 23: 2898–2908.
Acknowledgements
We thank Dr. G Corte (IST) for sharing lab facilities. Part of the studies has been conducted in the laboratories and facilities of the Centro Biotecnologie Avanzate (CBA, Genova, Italy). This work has been partly supported by grants from Italian ISS (527B/2B/6), Associazione Italiana per la Ricerca sul Cancro (AIRC), and CIPE 2007 (Regione Liguria, RNA Technology) to RG; from the Canadian Institute of Health Research (CIHR, MOP89798) to IEG; ISS (526D/39) and Fondazione Telethon (no. GGP04012) to PB. MT is a recipient of a Fondazione Telethon fellowship. PB is a recipient of a Senior Scholar Consultancy grant from AICF.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Cossu
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Gherzi, R., Trabucchi, M., Ponassi, M. et al. Akt2-mediated phosphorylation of Pitx2 controls Ccnd1 mRNA decay during muscle cell differentiation. Cell Death Differ 17, 975–983 (2010). https://doi.org/10.1038/cdd.2009.194
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2009.194
Keywords
This article is cited by
-
SHORT Syndrome: an Update on Pathogenesis and Clinical Spectrum
Current Diabetes Reports (2022)
-
MiR-17 and miR-19 cooperatively promote skeletal muscle cell differentiation
Cellular and Molecular Life Sciences (2019)
-
Glycosyltransferase ST6GAL1 contributes to the regulation of pluripotency in human pluripotent stem cells
Scientific Reports (2015)
-
CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay
Scientific Reports (2012)
-
PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis
Cell Death & Differentiation (2012)