Abstract
Multiple myeloma (MM) is an incurable haematological malignancy characterised by the proliferation of mature antibody-secreting plasma B cells in the bone marrow. MM can arise from initiating translocations, of which the musculoaponeurotic fibrosarcoma (MAF) family is implicated in ∼5%. MMs bearing Maf translocations are of poor prognosis. These translocations are associated with elevated Maf expression, including c-MAF, MAFB and MAFA, and with t(14;16) and t(14;20) translocations, involving c-MAF and MAFB, respectively. c-MAF is also overexpressed in MM through MEK/ERK activation, bringing the number of MMs driven by the deregulation of a Maf gene close to 50%. Here we demonstrate that MAFB and c-MAF are phosphorylated by the Ser/Thr kinase GSK3 in human MM cell lines. We show that LiCl-induced GSK3 inhibition targets these phosphorylations and specifically decreases proliferation and colony formation of Maf-expressing MM cell lines. Interestingly, bortezomib induced stabilisation of Maf phosphorylation, an observation that could explain, at least partially, the low efficacy of bortezomib for patients carrying Maf translocations. Thus, GSK3 inhibition could represent a new therapeutic approach for these patients.
Similar content being viewed by others
Introduction
Multiple myeloma (MM) is an incurable B-cell neoplasm of the bone marrow with a complex array of clinical manifestations.1, 2, 3, 4, 5 MM is further divided into seven disease subtypes based on molecular heterogeneity.6, 7 Of these, one subtype corresponds to the recurrent translocations t(14;16) and t(14;20) involving different musculoaponeurotic fibrosarcoma (Maf) genes juxtaposed to immunoglobulin heavy chain enhancer elements, leading to their aberrant overexpression. Although still debated due to its relatively low prevalence, this group was shown to be associated with poor overall survival in MM.4, 5, 8, 9, 10, 11, 12
Maf oncoproteins are basic leucine zipper transcription factors belonging to the AP-1 super family.13 The Maf family is composed of seven members that can be classified into two subfamilies, the large and small Maf genes. The large Maf members, MAFA, MAFB, c-MAF and NRL, differ from the small Maf members (MAFF, MAFG and MAFK) by the presence of a transactivation domain in their amino terminus.13 We and others demonstrated that large Maf proteins display strong transforming activity in chicken embryo fibroblasts, with MAFA being the most potent oncoprotein.14, 15, 16 We showed that MAFA-transforming activity in chicken embryo fibroblasts requires its phosphorylation by GSK3.17 Indeed, MafA is sequentially phosphorylated by GSK3 on residues S61, T57, T53 and S49.17, 18 At the molecular level, these phosphorylations coupled two antagonistic processes. GSK3-mediated MAFA phosphorylation increased its transactivation activity but also induced its degradation.17
Approximately 5% of MMs bear Maf translocations that are associated with elevated Maf expression including c-MAF, MAFB and MAFA.7, 19, 20, 21 The t(14;16) and t(14;20) translocations involving c-MAF and MAFB, respectively, are much more prevalent, with less than 0.5% of MMs displaying MAFA translocations.1, 4, 5, 7, 13 In ∼50% of MM patient samples lacking Maf translocations, c-MAF expression was observed at levels above those in normal plasma cells.22, 23 Maf overexpression has a causative role in MM22 by regulating cyclin D2, integrin β7, CCR1, ARK5 and DEPTOR expression, all of which have an important role in the pathogenesis of MM.22, 24, 25, 26, 27 These findings have been recently validated in a transgenic model, which develops B-cell lymphomas with features resembling MM-like disease highlighting Maf proteins as potential therapeutic targets in MM.28 The latest advances in MM treatment include combination of immunomodulatory agents with the proteasome inhibitor drug, bortezomib.5, 10, 29, 30 However, bortezomib appears not to be beneficial for patients carrying a Maf translocation.5, 8, 29 Therefore, there is an urgent need to identify additional therapeutic agents for the treatment of these patients with a poor outcome.
In the present study, we demonstrate that MAFB and c-MAF, the most frequently deregulated Maf in MM, are phosphorylated by GSK3, which mediates their degradation. Pharmacological inhibition of GSK3 targets these phosphorylations and leads to the decrease of MM cell proliferation and colony formation. This study provides the basis for further exploring GSK3 inhibition by lithium chloride (LiCl) in Maf-driven MMs in a clinical setting.
Materials and methods
Plasmid construction
Human MAFB and c-MAF wild type (WT) and mutated (4A) cDNAs were introduced in the pcDNA3 vector into the BamHI/EcoRI and HindIII/EcoRI restriction sites31 using PCR strategies.
Cell culture and transfection
Human MM cell lines (HMCLs) RPMI8226 (RPMI), H929 (ATCC), JJN3, KMS12PE (KMS12), LP-1, L363, OPM2 (DSMZ), KMS28BM, KMS28PE and KMM1 (Japanese health sciences foundation) were cultured in RPMI1640 medium supplemented with FBS. KMS12 cells were cultured in RPMI1640–20% FBS and JJN3 in 40% DMEM, 40% IMDM and 20% FBS. For transfection, HEK293T cells were cultured in DMEM medium supplemented with 10% FBS and transfected by calcium phosphate method.
In vitro proliferation assays
Proliferation was assessed under serum-starving conditions (0.2% FBS), in the presence of the GSK3 inhibitor LiCl (Sigma, Saint-Quentin Fallavier, France). Cells were treated on day 1 with 10 mM LiCl followed by daily treatment with 2.5 mM LiCl. Cells were seeded in duplicate at a density of 2 × 105 cells per 60-mm tissue culture dish and cultured for 7 days. Live cell counts were performed on days 1, 2, 3, 5 and 7.
Half-life studies, MG132, bortezomib treatment and western blotting
Total cell extracts were prepared in TNTSE lysis buffer (140 mM NaCl, 20 mM Tris pH 8, 2 mM EDTA, NP40, 1% triton, 0.2% SDS, 20 μg/ml aprotonin, 1 mM AEBSF, 10 mM NaF, 1 mM DTT, 25 mM β-glycerophosphtae and 1 mM orthovanadate). Nuclear protein extracts were prepared according to manufacturer’s guidelines (Thermo Scientific, Courtaboeuf, France). Cells were treated with 20 mM LiCl (Sigma) and the following pharmacological inhibitors: 10 μM SB216763 (Sigma), 10 μM MG132 (Sigma) and 100 nM Bortezomib (Cell Signaling Technology, Danvers, MA, USA). For bortezomib and MG132 treatment, cells were treated or not with LiCl for 1 h followed by MG132 or bortezomib treatment for 3 h. For half-life studies, cells were treated with 20 μg/ml cycloheximide (Sigma) for 30 min, 1, 2, 4 and 6 h time points in the presence or absence of LiCl.
Protein extracts were subjected to SDS-PAGE, transferred to Immobilon-P membranes (Millipore, Molsheim, France) and probed with anti-MafA (Abcam, Paris, France, 1/2000), anti-MAFB (Santa Cruz, Heidelberg, Germany, sc-10022 p20, 1/2000), anti-c-MAF (Santa Cruz, M153, 1/2000), anti-SAM68 (Santa Cruz, 1/5000), anti-β-actin (Sigma, 1/50000) and rabbit polyclonal T53-T57 phospho-specific (Rocques et al.,17 1/2000) antibodies. After three washes in PBST buffer, membranes were incubated with the corresponding secondary antibody. Membranes were processed using chemiluminescence. Band intensities were measured using the G:box (Syngene, Saint Quentin en Yvelines, France) and normalised to β-actin.
Colony formation assay (Soft agar cell culture)
A mixture of 2 ml of pre-warmed (37 °C) RPMI1640 medium supplemented with 10% FBS and 2 ml pre-warmed (42 °C) 0.66% Bacto Agar Select/plate (Sigma) (0.33% final agar) was mixed with a 2 × 105 cell suspension and seeded over a 0.66% agar/full medium pre-layer (7 ml) in a 60-mm dish. The cells were allowed to grow in a humidified 37 °C incubator with 5% CO2 for 3–4 weeks. Colony formation assays were assessed in the presence or absence of 5 mM LiCl, 10 μm GSK3 inhibitor VIII or 5 μm SB216763 (Sigma). Plates were scanned and the colony numbers were quantified using the ImageJ software.
RNA preparation and quantitative RT-PCR analysis
Total RNA extractions (Qiagen, Courtaboeuf Cedex, France) and reverse transcription (Invitrogen, Cergy Pontoise, France) were performed according to manufacturer’s recommendations. Quantitative real-time PCR (qPCR) analyses were performed using the SYBR Green master mix (Life Technologies, Courtaboeuf Cedex, France). Reactions were carried out in an iCycler Thermal Cycler (Bio-Rad Laboratories, Marnes-la-Coquette, France) for 40 cycles (95 °C for 15 s/60 °C for 1 min) after an initial 10-min incubation at 95 °C. mRNA levels were normalised to levels of HPRT mRNA. All primer sequences are listed in Table 1.
Statistical analyses
Two-sided, equal variance t-tests were used for colony formation and qPCR assays. Data from proliferation curves between Li-treated and untreated cell lines were analysed using non-parametric Wilcoxon rank sum tests. All statistical analyses were performed using StatEL software (ad Science, Paris, France) and a P-value of <0.05 was considered statistically significant.
Results
GSK3 phosphorylates MAFB and c-MAF proteins
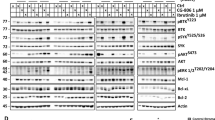
We previously demonstrated that MAFA is sequentially phosphorylated on residues Serine 61 (S61), Threonine 57, 53, 49 (T57, T53 and S49) by GSK3.17 These putative GSK3 phosphorylation sites are highly conserved amongst the large Maf proteins; MAFB and c-MAF (Figure 1a). In the current study, we focused on c-MAF and MAFB, the deregulation of which is the most prevalent in MM.
GSK3 phosphorylates MAFB and c-MAF proteins. (a) Amino acid sequence comparison of the large human Maf proteins (MAFA, MAFB and c-MAF). Putative GSK3 phosphorylation sites that are conserved among the different Maf proteins are highlighted in yellow. In the mutant unphosphorylatable form (4A), these putative GSK3 phosphorylation sites are mutated into alanine A. 293T cells were transfected using a pcDNA3-derived vector encoding the wild type (WT) and 4A mutants of human c-MAF (b), MAFB (c) and MAFA (d). GSK3 inhibitor treatment was done with 20 mM LiCl or 10 μM SB216763 (SB), on c-MAF, MAFB and MAFA, which demonstrates a comparable change for WT forms but had no effect on the 4A mutants. Total cell extracts were assessed by western blotting using the corresponding Maf antibodies, T53-57 phospho-specific MAFA (p-MAF) and β-actin antibodies. For MAFB (c), owing to a contaminating band in the close proximity of MAFB, nuclear extracts were used. Normalisation was carried out using an anti-Sam68 antibody. The p-MAF antibody cross-reacts with the phosphorylated forms of human c-MAF, MAFB and MAFA. NS, non-specific; P, phosphorylated forms.
To investigate whether MAFB and c-MAF are phosphorylated by GSK3 on the corresponding residues, we generated constructs expressing the WT phosphorylatable forms and the mutant unphosphorylatable forms (4A) where the four putative GSK3 phosphorylation sites were mutated into alanine (Figure 1a). HEK293T cells were transfected with pcDNA3-derived vectors encoding the WT and 4A mutants of c-MAF, MAFB and MAFA as a control, and cell lysates were analysed by western blotting. Mutation of the four putative GSK3 phosphorylation sites profoundly altered the migration pattern of c-MAF (Figure 1b), MAFB (Figure 1c) and MAFA (Figure 1d). Treatment with LiCl or SB216763 (SB), two GSK3 inhibitors, led to a comparable change for WT forms but had no effect on the 4A mutants. The migration patterns of the 4A mutants are similar to their corresponding WT forms on treatment with GSK3 inhibitors, suggesting that these residues are phosphorylated by GSK3. To confirm this phosphorylation, we made use of a previously described phospho-specific antibody that recognises T53 and T57 of chicken MAFA when dually phosphorylated (p-MAF Ab).17 This antibody also cross-reacts with the corresponding residues on MAFB (T58 and T62) and c-MAF (T58 and T62) when phosphorylated. Accordingly, the WT forms of human MAFA, MAFB and c-MAF but not the different 4A mutants were recognised by this antibody (Figure 1, middle panel). Treatment with GSK3 inhibitors prevented the detection of the WT proteins. Taken together, these results demonstrate that T58 and T62 of MAFB and c-MAF are phosphorylated by GSK3 in HEK293T. We next addressed whether c-MAF and MAFB are phosphorylated in MM.
GSK3 phosphorylates MAFB and c-MAF in MM
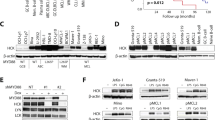
We undertook RT-qPCR to identify c-MAF- or MAFB-expressing HMCLs (Figure 2a). Western blot analysis was performed on extracts prepared from different HMCLs, expressing MAFB (L363 and OPM2) and c-MAF ((RPMI8226, H929, LP-1 and JJN3 KMM1) and from cell lines with no endogenous expression of Maf proteins (KMS12PE (KMS12), KMS28PE and KMS28BM) as negative controls, using the p-Maf Ab (Figures 2c–f). MAFB and c-MAF phosphorylation on T58 and T62 was detected in each cell line expressing these transcription factors (Figures 2c and d). As all HMCLs express GSK3α and GSK3β (Supplementary Figure S2A and S2B), we assessed whether these phosphorylations were mediated by GSK3. MM cell lines were treated with LiCl, a GSK3 inhibitor. The efficacy of this treatment was assessed through β-catenin accumulation, a known target of GSK3 (Supplementary Figure S2C). LiCl treatment led to the inhibition of MAFB and c-MAF phosphorylation (Figures 2e and f and Supplementary Figures S2D and S2E). This result demonstrates that Maf proteins are phosphorylated by GSK3 in HMCLs.
MAFB and c-MAF are phosphorylated by GSK3 in MM. Relative c-MAF (a) and MAFB (b) expression in MM cell lines (HMCLs) was determined by RT-qPCR. HMCL’s nuclear extracts were analysed by western blot using c-MAF (c) and MAFB (d) antibodies, p-MAF and SAM68 antibodies. c-MAF (e) and MAFB (f) expressing cell lines were treated with LiCl as indicated. Anti-c-MAF, anti-MAFB, p-MAF and SAM68 western blots were carried out on nuclear extracts. NS, non-specific; P, phosphorylated forms.
GSK3-mediated phosphorylation induces MAFB and c-MAF degradation in MM
We next assessed whether these phosphorylations alter Maf stability in MM. Treatment with the proteasome inhibitors MG132 and bortezomib lead to the stabilisation of the slower migrating bands of both c-MAF and MAFB (Figures 3a–d, upper panel) that correspond to the phosphorylated forms. This result was confirmed using the p-Maf Ab (Figures 3a–d, middle panel). T58 and T62 phosphorylation of MAFB and c-MAF increased on MG132 and bortezomib treatment, whereas disappearing on LiCl treatment, confirming that GSK3-mediated Maf phosphorylation leads to their degradation through the proteasome. To further support this conclusion, L363 (MAFB expressing) and RPMI (c-MAF expressing) cell lines were used for half-life studies (Figures 3e and f). Cycloheximide treatment demonstrated a shorter half-life for the phosphorylated forms of both c-MAF and MAFB and a much longer half-life in dephosphoryated forms induced by LiCl treatment. These results were confirmed on the WT and 4A forms of c-MAF and MAFB in 293T cells (Supplementary Figure S1). Taken together, these results show that GSK3-mediated MAFB and c-MAF phosphorylation leads to their degradation through a proteasome-dependent pathway in MM.
LiCl-mediated GSK3 inhibition leads to MAFB and c-MAF degradation and decreases DEPTOR expression. RPMI (a–c) and L363 (b–d) cells were treated or not with LiCl for 1 h and then with MG132 (MG) (a and b) or bortezomib (Btz) (c–d) for 3 h. Anti-c-MAF, MAFB, p-MAF and SAM68 western blots were carried out on nuclear extracts. RPMI (e) and L363 (f) cells were treated or not treated (w/o) with LiCl followed by treatment with 20 μg/ml cycloheximide (Chx) for the indicated time points. Nuclear extracts were analysed by western blots using c-MAF, MAFB, p-Maf and SAM68 antibodies. Band intensities were measured, normalised to SAM68 and plotted as a percentage of the initial band intensity (right panels). (g) The Relative DEPTOR expression in different HMCLs treated with LiCl (as indicated) was determined by RT-qPCR. Expression was normalised to HPRT. NS, non-specific; P, phosphorylated forms. DEPTOR expression was specifically decreased upon LiCl treatment in MAFB (L363 P=0.07, OPM2 P=NS) and c-MAF-expressing cells (RPMI P<0.05), but not in non-MAF-expressing cells (P=NS).
The expression of the MAF target gene DEPTOR is sensitive to LiCl treatment
We investigated whether inhibition of GSK3 could lead to a modulation of Maf target gene expression in HMCLs. We examined the effect of GSK3 inhibition on LiCl treatment on the expression level of different Maf target genes. No changes were detected for ITGB7 and CCND2 expression (data not shown). In contrast, DEPTOR expression was specifically decreased on LiCl treatment in MAFB- and c-MAF-expressing cells, but not in non-Maf-expressing cells (Figure 3e). Therefore, phosphorylation of c-MAF and MAFB appears to be correlated with an induction of DEPTOR expression, although their stability is decreased.
LiCl treatment decreases cell proliferation and colony formation in Maf-expressing HMCLs
As LiCl was able to target Maf phosphorylation in MM, we next investigated whether Maf-expressing HMCLs were sensitive to this GSK3 inhibitor. We first assessed whether LiCl treatment could affect cell proliferation. For this purpose, we performed growth curve analyses under low serum conditions on different HMCLs expressing (RPMI, L363 and OPM2) or not expressing (KMS12, KMS28BM and KMS28PE) Maf proteins in the presence or absence of LiCl. As shown in Figure 4, a significant correlation was observed between Maf-expressing cells and sensitivity to LiCl. Interestingly, the rate of proliferation of Maf-negative cell lines was significantly less affected by LiCl than c-MAF- or MAFB-expressing cell lines (Figures 4a and b). To gain further insight into the effect of LiCl on Maf-mediated transformation in MM, we tested the effect of LiCl on colony formation under semi-solid conditions (Figure 4c). We seeded L363, RPMI, LP-1 and KMS12 on semi-solid medium in the presence and absence of LiCl. Although LiCl significantly decreased colony formation in Maf-expressing cell lines, L363, LP-1 and RPMI, it had no effect on Maf-negative cell lines (KMS12). These results were confirmed using two structurally unrelated GSK3 inhibitors (GSK3 inhibitor VIII and SB216763) (Supplementary Figure S2 and S3). In conclusion, Maf-expressing HMCLs display sensitivity to LiCl.
LiCl treatment decreases cell proliferation and colony formation in Maf-expressing MM cells. Maf-expressing cell lines (RPMI, LP-1, L363 and OPM2) are indicated in blue and non-expressing cell lines (KMS12, KMS28PE and KMS28BM) in black. (a) These cell lines were treated without (w/o) and with 10 mM LiCl. Proliferation assays were conducted and live cell counts were performed in triplicate, on days, 0, 1, 2, 3, 5 and 7. (b) To directly assess the LiCl sensitivity on the rate of HMCLs proliferation, the results obtained in a were graphed for each cell line as the number of cells in LiCl-treated culture relative to that of the non-treated condition. The average rate of proliferation of Maf-negative cell lines was significantly less affected by LiCl than c-MAF- or MAFB-expressing cell lines (P<0.05). (c) Phase contrast images of soft agar assays without (w/o) and with 5 mM LiCl treatment. Plates were scanned, and the colony numbers were quantified using ImageJ and plotted for each cell line. Medium containing or not LiCl was renewed once a week for 3–4 weeks. LiCl significantly decreased colony formation in Maf-expressing cell lines; L363, LP-1 and RPMI (P<0.05), it had no effect on Maf-negative cells (KMS12, P=NS).
Discussion
Owing to its negative effect on β-catenin stability, GSK3 is usually thought to have an inhibitory role in tumorigenic processes. However, several studies reported a positive role for GSK3 in cancer.32, 33, 34 For example, it represents a potential therapeutic target in mixed-lineage leukaemia.35, 36 Recently, it was also demonstrated to have a pro-survival function in MM by permitting the activation of the non-canonical NFκB pathway.37 In this study, we demonstrate that MAFB and c-MAF are phosphorylated by GSK3 on residues T62 and T58 in MM cell lines. At the biochemical level, our results are compatible with a model where phosphorylation of c-MAF and MAFB in MM may trigger two antagonist effects, namely their destabilisation and an increase of their transactivation ability at least for a subset of target genes, as previously demonstrated for MAFA in Chicken embryo fibroblasts.17 This raises the possibility that degradation could be required for Maf transactivation, as demonstrated for other transcription factors.38 We show that these phosphorylations can be targeted by the GSK3 inhibitor, lithium, a therapeutic agent already approved for patients suffering from mood disorders.39, 40 Importantly, proliferation and colony formation of Maf-expressing cells are particularly sensitive to LiCl-mediated GSK3 inhibition. Our results suggest that the maintenance of these phosphorylations is essential for MAFB- and c-MAF-transforming activity in MM. Interestingly, proteasome inhibition by bortezomib leads to the stabilisation of Maf phosphorylation and overall Maf accumulation, an observation that could explain, at least partially, the low efficacy of bortezomib for patients carrying Maf translocations. In conclusion, aside from its role in non-canonical NFκB signalling, GSK3 also controls Maf phosphorylation, which could constitute a therapeutic target in MM. Our study suggests that patients bearing a Maf-driven MM could particularly benefit from therapies targeting GSK3.
References
Chng WJ, Glebov O, Bergsagel PL, Kuehl WM . Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol 2007; 20: 571–596.
Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC . Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer 2007; 7: 585–598.
Kuehl WM, Bergsagel PL . Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer 2002; 2: 175–187.
Morgan GJ, Walker BA, Davies FE . The genetic architecture of multiple myeloma. Nat Rev Cancer 2012; 12: 335–348.
Chesi M, Bergsagel PL . Many multiple myelomas: making more of the molecular mayhem. Hematol Am Soc Hematol Educ Program 2011; 2011: 344–353.
Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S et al. The molecular classification of multiple myeloma. Blood 2006; 108: 2020–2028.
Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia 2009; 23: 2210–2221.
Bergsagel PL, Mateos MV, Gutierrez NC, Rajkumar SV, San Miguel JF . Improving overall survival and overcoming adverse prognosis in the treatment of cytogenetically high-risk multiple myeloma. Blood 2013; 121: 884–892.
Munshi NC, Anderson KC, Bergsagel PL, Shaughnessy J, Palumbo A, Durie B et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood 2011; 117: 4696–4700.
Rajkumar SV . Treatment of multiple myeloma. Nat Rev Clin Oncol 2011; 8: 479–491.
Rajkumar SV . Multiple myeloma: 2013 update on diagnosis, risk-stratification, and management. Am J Hematol 2013; 88: 225–235.
Avet-Loiseau H, Malard F, Campion L, Magrangeas F, Sebban C, Lioure B et al. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood 2011; 117: 2009–2011.
Eychene A, Rocques N, Pouponnot C . A new MAFia in cancer. Nat Rev Cancer 2008; 8: 683–693.
Pouponnot C, Sii-Felice K, Hmitou I, Rocques N, Lecoin L, Druillennec S et al. Cell context reveals a dual role for Maf in oncogenesis. Oncogene 2006; 25: 1299–1310.
Kataoka K, Shioda S, Yoshitomo-Nakagawa K, Handa H, Nishizawa M . Maf and Jun nuclear oncoproteins share downstream target genes for inducing cell transformation. J Biol Chem 2001; 276: 36849–36856.
Nishizawa M, Kataoka K, Vogt PK . MafA has strong cell transforming ability but is a weak transactivator. Oncogene 2003; 22: 7882–7890.
Rocques N, Abou Zeid N, Sii-Felice K, Lecoin L, Felder-Schmittbuhl MP, Eychene A et al. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol Cell 2007; 28: 584–597.
Han SI, Aramata S, Yasuda K, Kataoka K . MafA stability in pancreatic beta cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol Cell Biol 2007; 27: 6593–6605.
Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood 1998; 91: 4457–4463.
Hanamura I, Iida S, Akano Y, Hayami Y, Kato M, Miura K et al. Ectopic expression of MAFB gene in human myeloma cells carrying (14;20)(q32;q11) chromosomal translocations. Jpn J Cancer Res 2001; 92: 638–644.
Boersma-Vreugdenhil GR, Kuipers J, Van Stralen E, Peeters T, Michaux L, Hagemeijer A et al. The recurrent translocation t(14;20)(q32;q12) in multiple myeloma results in aberrant expression of MAFB: a molecular and genetic analysis of the chromosomal breakpoint. Br J Haematol 2004; 126: 355–363.
Hurt EM, Wiestner A, Rosenwald A, Shaffer AL, Campo E, Grogan T et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 2004; 5: 191–199.
Annunziata CM, Hernandez L, Davis RE, Zingone A, Lamy L, Lam LT et al. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood 2011; 117: 2396–2404.
Suzuki A, Iida S, Kato-Uranishi M, Tajima E, Zhan F, Hanamura I et al. ARK5 is transcriptionally regulated by the Large-MAF family and mediates IGF-1-induced cell invasion in multiple myeloma: ARK5 as a new molecular determinant of malignant multiple myeloma. Oncogene 2005; 24: 6936–6944.
Morito N, Yoh K, Fujioka Y, Nakano T, Shimohata H, Hashimoto Y et al. Overexpression of c-Maf contributes to T-cell lymphoma in both mice and human. Cancer Res 2006; 66: 812–819.
Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009; 137: 873–886.
van Stralen E, van de Wetering M, Agnelli L, Neri A, Clevers HC, Bast BJ . Identification of primary MAFB target genes in multiple myeloma. Exp Hematol 2009; 37: 78–86.
Morito N, Yoh K, Maeda A, Nakano T, Fujita A, Kusakabe M et al. A novel transgenic mouse model of the human multiple myeloma chromosomal translocation t(14;16)(q32;q23). Cancer Res 2011; 71: 339–348.
Kuehl WM, Bergsagel PL . Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest 2012; 122: 3456–3463.
Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K . Latest advances and current challenges in the treatment of multiple myeloma. Nat Rev Clin Oncol 2012; 9: 135–143.
Benkhelifa S, Provot S, Lecoq O, Pouponnot C, Calothy G, Felder-Schmittbuhl MP . mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene 1998; 17: 247–254.
Patel S, Woodgett J . Glycogen synthase kinase-3 and cancer: good cop, bad cop? Cancer Cell 2008; 14: 351–353.
Billadeau DD . Primers on molecular pathways. The glycogen synthase kinase-3beta. Pancreatology 2007; 7: 398–402.
Ougolkov AV, Billadeau DD . Targeting GSK-3: a promising approach for cancer therapy? Future Oncol 2006; 2: 91–100.
Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML . Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature 2008; 455: 1205–1209.
Wang Z, Iwasaki M, Ficara F, Lin C, Matheny C, Wong SH et al. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell 2010; 17: 597–608.
Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O'Connor O et al. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol 2012; 14: 375–385.
Geng F, Wenzel S, Tansey WP . Ubiquitin and proteasomes in transcription. Annu Rev Biochem 2012; 81: 177–201.
Doble BW, Woodgett JR . GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci 2003; 116: 1175–1186.
Li X, Jope RS . Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology 2010; 35: 2143–2154.
Acknowledgements
We thank Christophe Alberti and Elodie Belloir for technical assistance. We are also grateful to Drs Dominique Dusmenil, Isabelle Dusanter and Evelyne Lauret for the transfer of various technologies and helpful advice. This work was funded by the Fondation ARC (grant number #5089) and Ligue contre le cancer (grant numbers CV/CB/055-10 and NM/CP/M16649). NH received fellowships from Fondation ARC and Institut Curie.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Blood Cancer Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Herath, N., Rocques, N., Garancher, A. et al. GSK3-mediated MAF phosphorylation in multiple myeloma as a potential therapeutic target. Blood Cancer Journal 4, e175 (2014). https://doi.org/10.1038/bcj.2013.67
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2013.67
Keywords
This article is cited by
-
The role and regulation of Maf proteins in cancer
Biomarker Research (2023)
-
Multicentric Carpotarsal Osteolysis: a Contemporary Perspective on the Unique Skeletal Phenotype
Current Osteoporosis Reports (2023)
-
circFBXO7/miR-96-5p/MTSS1 axis is an important regulator in the Wnt signaling pathway in ovarian cancer
Molecular Cancer (2022)
-
The proliferative history shapes the DNA methylome of B-cell tumors and predicts clinical outcome
Nature Cancer (2020)
-
MAFb protein confers intrinsic resistance to proteasome inhibitors in multiple myeloma
BMC Cancer (2018)