Abstract
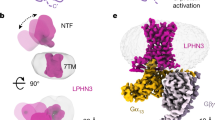
Adhesion G protein-coupled receptors (aGPCRs) constitute an evolutionarily ancient family of receptors that often undergo autoproteolysis to produce α and β subunits1,2,3. A tethered agonism mediated by the ‘Stachel sequence’ of the β subunit has been proposed to have central roles in aGPCR activation4,5,6. Here we present three cryo-electron microscopy structures of aGPCRs coupled to the Gs heterotrimer. Two of these aGPCRs are activated by tethered Stachel sequences—the ADGRG2-β–Gs complex and the ADGRG4-β–Gs complex (in which β indicates the β subunit of the aGPCR)—and the other is the full-length ADGRG2 in complex with the exogenous ADGRG2 Stachel-sequence-derived peptide agonist IP15 (ADGRG2(FL)–IP15–Gs). The Stachel sequences of both ADGRG2-β and ADGRG4-β assume a U shape and insert deeply into the seven-transmembrane bundles. Constituting the FXφφφXφ motif (in which φ represents a hydrophobic residue), five residues of ADGRG2-β or ADGRG4-β extend like fingers to mediate binding to the seven-transmembrane domain and activation of the receptor. The structure of the ADGRG2(FL)–IP15–Gs complex reveals the structural basis for the improved binding affinity of IP15 compared with VPM–p15 and indicates that rational design of peptidic agonists could be achieved by exploiting aGPCR-β structures. By converting the ‘finger residues’ to acidic residues, we develop a method to generate peptidic antagonists towards several aGPCRs. Collectively, our study provides structural and biochemical insights into the tethered activation mechanism of aGPCRs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data generated in this study are included in the main text or the Supplementary Information. The atomic coordinates and the cryo-EM density maps have been deposited in the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMDB) under accession codes 7WUQ and EMD-32838 for the ADGRG2-β–Gs complex; 7WUJ and EMD-32837 for the ADGRG4-β–Gs complex; and 7WUI and EMD-32836 for the ADGRG2(FL)–IP15–Gs complex, respectively. All other data are available upon request to the corresponding authors.
References
Bassilana, F., Nash, M. & Ludwig, M. G. Adhesion G protein-coupled receptors: opportunities for drug discovery. Nat. Rev. Drug Discov. 18, 869–884 (2019).
Bondarev, A. D. et al. Opportunities and challenges for drug discovery in modulating adhesion G protein-coupled receptor (GPCR) functions. Expert Opin. Drug Discov. 15, 1291–1307 (2020).
Hamann, J. et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 67, 338–367 (2015).
Liebscher, I. & Schoneberg, T. Tethered agonism: a common activation mechanism of adhesion GPCRs. Handb. Exp. Pharmacol. 234, 111–125 (2016).
Stoveken, H. M., Hajduczok, A. G., Xu, L. & Tall, G. G. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl Acad. Sci. USA 112, 6194–6199 (2015).
Beliu, G. et al. Tethered agonist exposure in intact adhesion/class B2 GPCRs through intrinsic structural flexibility of the GAIN domain. Mol. Cell 81, 905–921 (2021).
Purcell, R. H. & Hall, R. A. Adhesion G protein-coupled receptors as drug targets. Annu. Rev. Pharmacol. Toxicol. 58, 429–449 (2018).
Paavola, K. J. & Hall, R. A. Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 82, 777–783 (2012).
Arac, D. et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 31, 1364–1378 (2012).
Krasnoperov, V. et al. Post-translational proteolytic processing of the calcium-independent receptor of α-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J. Biol. Chem. 277, 46518–46526 (2002).
Lin, H. H. et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 279, 31823–31832 (2004).
Kaur, B., Brat, D. J., Devi, N. S. & Van Meir, E. G. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene 24, 3632–3642 (2005).
Eubelen, M. et al. A molecular mechanism for Wnt ligand-specific signaling. Science 361, eaat1178 (2018).
Little, K. D., Hemler, M. E. & Stipp, C. S. Dynamic regulation of a GPCR–tetraspanin–G protein complex on intact cells: central role of CD81 in facilitating GPR56–Gαq/11 association. Mol. Biol. Cell 15, 2375–2387 (2004).
Ward, Y. et al. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 71, 7301–7311 (2011).
Kuffer, A. et al. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 536, 464–468 (2016).
Luo, R. et al. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl Acad. Sci. USA 108, 12925–12930 (2011).
Lv, X. et al. In vitro expression and analysis of the 826 human G protein-coupled receptors. Protein Cell 7, 325–337 (2016).
Stoveken, H. M., Larsen, S. D., Smrcka, A. V. & Tall, G. G. Gedunin- and khivorin-derivatives are small-molecule partial agonists for adhesion G protein-coupled receptors GPR56/ADGRG1 and GPR114/ADGRG5. Mol. Pharmacol. 93, 477–488 (2018).
Bianchi, E. et al. Control of oviductal fluid flow by the G-protein coupled receptor Adgrd1 is essential for murine embryo transit. Nat. Commun. 12, 1251 (2021).
Wilde, C. et al. The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. 30, 666–673 (2016).
Frenster, J. D. et al. Functional impact of intramolecular cleavage and dissociation of adhesion G protein-coupled receptor GPR133 (ADGRD1) on canonical signaling. J. Biol. Chem. 296, 100798 (2021).
Petersen, S. C. et al. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron 85, 755–769 (2015).
Scholz, N. et al. Mechano-dependent signaling by Latrophilin/CIRL quenches cAMP in proprioceptive neurons. eLife 6, e28360 (2017).
Yeung, J. et al. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc. Natl Acad. Sci. USA 117, 28275–28286 (2020).
Gupte, J. et al. Signaling property study of adhesion G-protein-coupled receptors. FEBS Lett. 586, 1214–1219 (2012).
Ping, Y. Q. et al. Structures of the glucocorticoid-bound adhesion receptor GPR97–Go complex. Nature 589, 620–626 (2021).
Liebscher, I. et al. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 9, 2018–2026 (2014).
Promel, S. et al. The GPS motif is a molecular switch for bimodal activities of adhesion class G protein-coupled receptors. Cell Rep. 2, 321–331 (2012).
Vizurraga, A., Adhikari, R., Yeung, J., Yu, M. & Tall, G. G. Mechanisms of adhesion G protein-coupled receptor activation. J. Biol. Chem. 295, 14065–14083 (2020).
Paavola, K. J., Sidik, H., Zuchero, J. B., Eckart, M. & Talbot, W. S. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci. Signal. 7, ra76 (2014).
de Groot, D. M. et al. Therapeutic antibody targeting of CD97 in experimental arthritis: the role of antigen expression, shedding, and internalization on the pharmacokinetics of anti-CD97 monoclonal antibody 1B2. J. Immunol. 183, 4127–4134 (2009).
Langenhan, T., Aust, G. & Hamann, J. Sticky signaling-adhesion class G protein-coupled receptors take the stage. Sci. Signal. 6, re3 (2013).
Zhang, D. et al. Function and therapeutic potential of G protein-coupled receptors in epididymis. Br. J. Pharmacol. 177, 5489–5508 (2020).
Zhang, D. L. et al. Gq activity- and beta-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. eLife 7, e33432 (2018).
Peeters, M. C. et al. Getting from A to B-exploring the activation motifs of the class B adhesion G protein-coupled receptor subfamily G member 4/GPR112. FASEB J. 30, 1836–1848 (2016).
Demberg, L. M., Rothemund, S., Schoneberg, T. & Liebscher, I. Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochem. Biophys. Res. Commun. 464, 743–747 (2015).
Sun, Y. et al. Optimization of a peptide ligand for the adhesion GPCR ADGRG2 provides a potent tool to explore receptor biology. J. Biol. Chem. 296, 100174 (2020).
Demberg, L. M. et al. Activation of adhesion G protein-coupled receptors: agonist specificity of Stachel sequence-derived peptides. J. Biol. Chem. 292, 4383–4394 (2017).
Vallon, M. & Essler, M. Proteolytically processed soluble tumor endothelial marker (TEM) 5 mediates endothelial cell survival during angiogenesis by linking integrin alphavβ3 to glycosaminoglycans. J. Biol. Chem. 281, 34179–34188 (2006).
Chiang, N. Y. et al. Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J. Cell Sci. 129, 2156–2169 (2016).
Chiang, N. Y. et al. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J. Biol. Chem. 286, 14215–14225 (2011).
Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl Acad. Sci. USA 110, 5211–5216 (2013).
de Graaf, C., Nijmeijer, S., Wolf, S. & Ernst, O. P. 7TM domain structure of adhesion GPCRs. Handb. Exp. Pharmacol. 234, 43–66 (2016).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Leon, K. et al. Structural basis for adhesion G protein-coupled receptor Gpr126 function. Nat. Commun. 11, 194 (2020).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Brown, K. et al. Epithelial Gpr116 regulates pulmonary alveolar homeostasis via Gq/11 signaling. JCI Insight 2, e93700 (2017).
Baxendale, S., Asad, A., Shahidan, N. O., Wiggin, G. R. & Whitfield, T. T. The adhesion GPCR Adgrg6 (Gpr126): insights from the zebrafish model. Genesis 59, e23417 (2021).
Hu, Q. X. et al. Constitutive Galphai coupling activity of very large G protein-coupled receptor 1 (VLGR1) and its regulation by PDZD7 protein. J. Biol. Chem. 289, 24215–24225 (2014).
Yang, F. et al. Structural basis of GPBAR activation and bile acid recognition. Nature 587, 499–504 (2020).
Wang, L. et al. Cryo-EM structure of the AVP-vasopressin receptor 2–Gs signaling complex. Cell Res. 31, 932–934 (2021).
Zhou, F. et al. Molecular basis of ligand recognition and activation of human V2 vasopressin receptor. Cell Res. 31, 929–931 (2021).
Bi, C. et al. A python script to design site-directed mutagenesis primers. Protein Sci. 29, 1054–1059 (2020).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Yang, F. et al. Structure, function and pharmacology of human itch receptor complexes. Nature 600, 164–169 (2021).
Bokoch, M. P. et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463, 108–112 (2010).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Liu, Q. et al. DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl (1)H-NMR probe. Nat. Commun. 11, 4857 (2020).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Metropolis, N. & Ulam, S. The Monte Carlo method. J. Am. Stat. Assoc. 44, 335–341 (1949).
He, Q. T. et al. Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2. Nat. Commun. 12, 2396 (2021).
Kumari, R., Kumar, R., Open Source Drug Discovery Consortium & Lynn, A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 54, 1951–1962 (2014).
Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFC1003600 to X.Y. and J.-P.S. and 2019YFA0904200 to J.-P.S. and P.X.); the National Natural Science Foundation of China (81773704 to J.-P.S., 92057121 to X.Y., 31971195 to P.X., 11922410 and 11834018 to L.Z.); the National Science Fund for Distinguished Young Scholars (81825022 to J.-P.S.); the National Science Fund for Excellent Young Scholars (81822008 to X.Y.); Shandong Provincial Natural Science Fund for Excellent Young Scholars (ZR2021YQ18 to P.X.); the Key Research and Development Program of Shandong Province (2021CXGC011105 to J.-P.S., 2021ZLGX02 to C.W. and J.-P.S.); the Key Research Project of the Natural Science Foundation of Beijing, China (Z200019 to J.-P.S.); the Major Fundamental Research Program of Natural Science Foundation of Shandong Province, China (ZR2021ZD18 to X.Y.); and Fundamental Research Funds for the Central Universities (2021JCG020 to J.-P.S. and P.X.). The cryo-EM data were collected at the cryo-EM facility of the National Center for Protein Science Shanghai with assistance from F.-F. Wang. The cryo-EM grids were prepared and optimized at the Instrumental Analysis Center of Xi’an Jiaotong University and the Electron Microscopy Center of Translational Medicine Core Facility of Advanced Medical Research Institute at Shandong University with assistance from L. Wang. The molecular simulations were performed on the HPC Cloud Platform of Shandong University. Cell-based cAMP GloSensor assays were performed using a PerkinElmer EnVision 2105 multimode plate reader at the Translational Medicine Core Facility of Advanced Medical Research Institute at Shandong University.
Author information
Authors and Affiliations
Contributions
X.Y. initiated the project for investigation of the tethered agonism of aGPCRs and ADGRG2. X.Y. and J.-P.S. supervised the overall project. L.K. and L.Z. supervised the determination of the three cryo-EM structures. X.Y., J.-P.S., P.X., S.G., X.W. and Q.-T.H. participated in data analysis and interpretation. Z.G. participated in the initial preparation of Sf9 cells for ADGRG2-β. P.X. and S.G. developed the procedure for the formation and purification of the ADGRG2-β–Gs and ADGRG2(FL)–IP15–Gs complexes under the supervision of J.-P.S. S.G., P.X. and S.-M.H. prepared ADGRG2-β–Gs, ADGRG4-β–Gs and ADGRG2(FL)–IP15–Gs complex samples for cryo-EM. L.G., P.X. and Y. Li prepared cryo-EM grids with the assistance of X.-N.T. L.K. performed the cryo-EM data collection. S.G., L.G., P.X. and S.-M.H. processed cryo-EM data with assistance from C.Z. and generated the cryo-EM density map under the supervision of L.Z. and L.K. Q.-T.H., P.X., S.G. and S.-M.H. built the models. P.X., Q.-T.H., S.G., X.W., C.Z. and X.-N.T. manually analysed the homology of Stachel sequences across the aGPCR subfamily under the supervision of J.-P.S. and X.Y. Q.-T.H. and C.Z. performed model simulations for the reset of aGPCR members according to the atomic coordinates of the ADGRG2-β–Gs complex using SWISS-MODEL software. J.-P.S., P.X. and Q.-T.H. designed all of the mutations related to the Stachel-sequence-dependent activation of aGPCR in ADGRG1, ADGRG3, ADGRG4 and ADGRG5. X.Y. and J.-P.S. conducted the design of antagonists. X.W., P.X. and S.-L.L. performed cell-based functional assays including the cAMP accumulation assay, cAMP inhibition assay, G protein dissociation assay and FlAsH BRET assay. H.L. performed the electrophysiological experiments with assistance from Z.Y. and C.W. X.W., Z.-S.Y., J.-Y.H. and C.-C.Y. generated all constructs used in this study. P.X., Q.-T.H., S.G., X.W. and L.G. prepared figures with assistance from Y.-N.Z. and Y. Lu X.Y. and J.-P.S. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Antony Boucard and Aashish Manglik for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 The presence of the individual ADGRG2-β and ADGRG4-β.
a, Several proposed activation models of aGPCRs that involve tethered agonisms. (I) Binding of soluble ligands (small molecules or extracellular proteins) induces conformational changes in the extracellular domain, which may facilitate the interaction between the Stachel sequence and the seven transmembrane (7TM) bundle of the aGPCR. (II) After autoproteolysis at the GPCR-proteolytic site (GPS, HLT), binding of matrix proteins or mechanical force may lead to dissociation of the α subunit from the β subunit, which exposes the tethered Stachel sequence and allows the binding of the Stachel sequence to the 7TM bundle. (III) Mechano-forces induce conformational changes in the extracellular domain, which allow the binding of the tethered Stachel sequence to the 7TM bundle. b, Constitutive activities of wild-type ADGRG2 (ADGRG2(FL)) and its truncated versions measured by the cAMP accumulation assay at similar receptor expression levels. The intracellular cAMP levels of ADGRG2-β and its mutants were measured at indicated expression levels as shown in 1c. Values are the mean ± s.e.m. of three independent experiments (n = 3) performed in triplicate. Comparisons between the ADGRG2(FL) and its truncated versions were determined by two-sided one-way ANOVA with Tukey’s test. ***P < 0.001, n.s., no significant difference. (P = 0.2419, 0.7303, 0.1913, 0.1368, 0.2735, 0.2553 from left to right for ADGRG2-β(HL) curve; P = 0.6623, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001 from left to right for ADGRG2(FL) curve; P = 0.0617, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001 from left to right for ADGRG2-β(∆S) curve). c, Expression levels of wild-type ADGRG2 (ADGRG2(FL)) and its truncated versions are shown by ELISA experiments when HEK293 cells were transiently transfected with indicated amounts of plasmids. Values are mean ± s.e.m. from three independent experiments (n = 3) performed in triplicate. Comparisons between the ADGRG2(FL) and its truncated versions were determined by two-sided one-way ANOVA with Tukey’s test. n.s., no statistical significance. (P = 0.9471, 0.3217, 0.9336, 0.5713, 0.2566, 0.6301 from left to right for ADGRG2-β(HL); P = 0.9536, 0.2560, 0.9763, 0.1235, 0.7507, 0.3880 from left to right for ADGRG2(FL); P = 0.1489, 0.8455, 0.7348, 0.2112, 0.4952, 0.8970 from left to right for ADGRG2-β(∆S)). d, Standard curves for quantification of Flag tagged-proteins and GFP-tagged proteins by western. Representative western blots from at least three independent experiments are shown. For uncropped gel scans, see Supplementary Fig. 1. The densitometry of the western blot bands of purified Flag-MRGPRX2 and GFP proteins were quantified using ImageJ and compared with input protein amounts using linear regression. Values are mean ± s.e.m. from three independent experiments (n = 3). e, f, Western blot analysis of ADGRG2 (e) and ADGRG4 (f) auto-proteolysis in HEK293 cells. Full gel scans are shown. Cells were transfected with wild-type or auto-proteolysis deficient ADGRG2/GAIN-ADGRG4-β (ADGRG2-H597A/T599A (AA) and GAIN-ADGRG4-β-H2720A/T2722A (AA)) with N terminal fused Flag and C terminal fused GFP tags. Whole-cell lysates were analysed by western blot using the Flag (α subunit; e-f, left panel) or GFP (β subunit; e-f, right panel) antibody. Cleaved ADGRG2/GAIN-ADGRG4 α subunit bands (~130/70 kDa) or β subunit bands (~60 kDa), and ADGRG2(FL)/GAIN-ADGRG4-β uncleaved bands (~180/120 kDa) or uncleavable ADGRG2-AA or GAIN-ADGRG4-β-AA bands (~180/120 kDa) are highlighted with blue/sky blue, red/orange, and green arrows, respectively. Representative blots from at least three independent experiments (n = 3) are shown. The percentage of auto-cleaved ADGRG2 (e, right panel) or GAIN-ADGRG4 (f, right panel) were obtained by dividing of the average densitometry of ADGRG2/ADGRG4 β subunit bands with the average densitometry of the ADGRG2-AA or GAIN-ADGRG4-β-AA bands. Values are mean ± s.e.m. from three independent experiments (n = 3). g, h, The co-immunoprecipitation analysis of the association between the α and β subunit of ADGRG2 (g) and of GAIN-ADGRG4-β (h) in HEK293 cells. β subunit of ADGRG2 or GAIN-ADGRG4-β were immune-precipitated from whole-cell lysates using anti-GFP antibody covalently linked sepharose beads. Full gel scans are shown. The immunoprecipitants were analysed by western blot and simultaneously co-stained with an antibody against the Flag (α subunit, g, h, left panel) and GFP (β subunit, g, h, middle panel). Cleaved ADGRG2/GAIN-GAIN-ADGRG4 α subunit bands (~130/70 kDa) or β subunit bands (~60 kDa), ADGRG2(FL)/GAIN-ADGRG4-β uncleaved bands (~180/120 kDa) or uncleavable ADGRG2-AA or GAIN-ADGRG4-β-AA bands (~180/120 kDa) are highlighted with blue/sky blue, red/orange, and green arrows, respectively. Representative blots from at least three independent experiments (n = 3) are shown. The association ratio of α and β subunit of ADGRG2 (g, right panel) or GAIN-ADGRG4-β (h, right panel) were obtained by dividing of the densitometry of α subunit bands with the β subunit bands, using the densitometry of the ADGRG2-AA or GAIN-ADGRG4-β-AA bands for normalization (Because we have subjected the same amount of the ADGRG2-AA/GAIN-ADGRG4-β-AA for western blot. The separation ratio between the α and β subunits were calculated by subtraction of 100% with the association α/β ratio). Values are mean ± s.e.m. from three independent experiments (n = 3). i, The whole cell Cl- current elicited by voltage steps between −100 mV and +100 mV in HEK293 cells overexpressing either CFTR and ADGRG2(FL), or CFTR and ADGRG2-AA, or CFTR alone. The whole-cell anion current recording and I-V analysis suggested that the CFTR current was significantly larger in wild-type ADGRG2- than in ADGRG2-AA-overexpressing HEK293 cells. j, Corresponding I-V curves of whole-cell Cl- current as shown in i. Values are the mean ± s.e.m. of six independent experiments (n = 6). k, Average current densities (pA/pF) recorded at 100 mV as shown in i. Values are the mean ± s.e.m. of six independent experiments (n = 6). ***P < 0.001; the anion current in HEK293 cells overexpressing CFTR and ADGRG2(FL) were compared with that in HEK293 cells overexpressing CFTR alone. ###P < 0.001; the anion current in HEK293 cells overexpressing CFTR and ADGRG2-AA were compared with that overexpressing CFTR and ADGRG2(FL). All data were analysed by two-sided one-way ANOVA with Tukey’s test. (P < 0.0001, 0.0008 from left to right). l, Effects of the GAIN-ADGRG4-β/GAIN-ADGRG4-β-AA on their constitutive activities were determined by measuring the basal cAMP levels. Values are the mean ± s.e.m. of three independent experiments (n = 3). Statistical differences between GAIN-ADGRG4-β and GAIN-ADGRG4-β-AA were determined by two-sided one-way ANOVA with Tukey’s test. ***P < 0.001; (P = 0.0002).
Extended Data Fig. 2 Mechanical-force-induced separation between the α and β subunit of ADGRG2, and CFTR current increase via ADGRG2.
a, The whole cell Cl- current elicited by voltage steps between −100 mV and +100 mV in HEK293 cells overexpressing either CFTR and ADGRG2(FL), or CFTR and ADGRG2-AA, or CFTR alone, after stimulation with mechano-forces (cells were treated with shaking at 60 rpm for 2 h as indicated by previous literature). Note that the nonciliated cells of efferent ductules enriched with ADGRG2 expression constantly withstood the pressure of fluids. We therefore examined the CFTR current in HEK293 cells overexpressing ADGRG2 and CFTR after stimulation with mechanical forces using a protocol from research on other aGPCRs. b, Corresponding I-V curves of whole-cell Cl- current as shown in a. Values are the mean ± s.e.m. of six independent experiments (n = 6). c, Average current densities (pA/pF) recorded at 100 mV as shown in b. Values are the mean ± s.e.m. of six independent experiments (n = 6). ***P < 0.001; *P < 0.05; n.s., no significant difference. HEK293 cells transfected with CFTR and ADGRG2(FL), or CFTR and ADGRG2-AA, or CFTR alone treated with or without shaking were compared. All data were analysed by two-sided one-way ANOVA with Tukey’s test. (P = 0.4908, <0.0001, 0.0215 from left to right). d, Box plots showed the increased current densities (pA/pF) recorded at 100 mV after mechano-forces administration as shown in b. In box plots, whiskers indicate the minimum and maximum values, the bounds of the box are 25th and 75th percentiles of the data, and the centre line indicates the mean. Values are the mean ± s.e.m. of six independent experiments (n = 6). Note that the CFTR current was increased after stimulation with mechanical forces, with a significantly larger increase in the wild-type ADGRG2 group than in the ADGRG2-AA group. ***P < 0.001; mechano-forces-stimulated increased current densities in CFTR and ADGRG2(FL) overexpressed cells compared with that in CFTR overexpressed cells. ###P < 0.001; mechano-forces-stimulated increased current densities in CFTR and ADGRG2-AA overexpressed cells compared with that in CFTR and ADGRG2(FL) overexpressed cells. All data were analysed by two-sided one-way ANOVA with Tukey’s test. (P <0.0001, <0.0001 from left to right). e, The auto-cleavage ratio of the ADGRG2 after application of mechano-force examined by western blot, as depicted in Extended Data Fig. 1e. Full gel scans are shown. Cells were transfected with wild-type or auto-proteolysis deficient ADGRG2 (ADGRG2-H597A/T599A (AA)) with Flag and GFP tags fused at their N- and C-terminus, respectively. Whole-cell lysates were analysed by western blot using anti-Flag (α subunit) or anti-GFP (β subunit) antibody. Western blot membranes were co-stained against the Flag (ADGRG2-α subunit, left panel) and GFP (ADGRG2 β subunit, middle panel). Cleaved ADGRG2 α subunit bands (~130 kDa), cleaved ADGRG2 β subunit bands (~60 kDa), and ADGRG2(FL) uncleaved bands (~180 kDa) or uncleavable ADGRG2-AA bands (~180 kDa) are highlighted with blue, red, and green arrows, respectively. Representative blots from at least three independent experiments (n=3) are shown. The percentage of auto-cleaved ADGRG2 (right panel) were obtained by dividing of the average densitometry of ADGRG2 β subunit bands with the average densitometry of the ADGRG2-AA bands. Values are mean ± s.e.m. from three independent experiments (n = 3). Notably, the sum of the average densitometries of the ADGRG2 β subunit bands and ADGRG2(FL) uncleaved bands were equal to the densitometry of the ADGRG2-AA bands. f, The co-immunoprecipitation analysis of the association between the α and β subunit of ADGRG2 after application of mechano-force, as depicted in Extended Data Fig. 1g. Full gel scans are shown. Representative blots from at least three independent experiments (n = 3) are shown. β subunit of ADGRG2 were immune-precipitated from whole-cell lysates using anti-GFP antibody covalently linked sepharose beads. The immune-precipitants were analysed by western blot and simultaneously co-stained with an antibody against the Flag (α subunit, left panel) and GFP (β subunit, middle panel). Cleaved ADGRG2 α subunit bands (~130 kDa), cleaved ADGRG2 β subunit bands (~60 kDa), ADGRG2(FL) uncleaved bands (~180 kDa) or uncleavable ADGRG2-AA bands (~180 kDa) are highlighted with blue, red, and green arrows, respectively. The association ratio of α and β subunit of ADGRG2 (right panel) were obtained by dividing of the densitometry of α subunit bands with the β subunit bands, using the densitometry of the ADGRG2-AA bands for normalization (Because we have subjected the same amount of the ADGRG2-AA/GAIN-ADGRG4-β-AA for western blot. The separation ratio between the α and β subunits were calculated by subtraction of 100% with the association α/β ratio.) Values are mean ± s.e.m. from three independent experiments (n = 3). Mechanical force stimulation accelerated the separation of the ADGRG2-β subunit from its α subunit because no detectable α subunit was found to be associated with the ADGRG2-β subunit by co-IP (cells were treated with shaking at 60 rpm for 2 h according to previous literature). We speculated that the mechanical force-induced separation ratio between the α and β subunits was 100%. Note that the mechanical forces in our experimental conditions probably accelerated the separation of the ADGRG2-β subunit from its α subunit because no detectable α subunit was found to be associated with the ADGRG2-β subunit by co-IP.
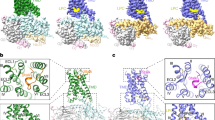
Extended Data Fig. 3 Cryo-EM density maps and validation of the ADGRG2-β–Gs, ADGRG4-β–Gs and ADGRG2(FL)–IP15–Gs complex structures.
a, Cryo-EM density map of the ADGRG2-β (left panel), ADGRG4-β (middle panel) and IP15-bound ADGRG2(FL) (right panel) in complex with the Gs heterotrimer. ADGRG2-β, green; ADGRG2-tethered Stachel sequence, blue; Gαs, yellow; Gβ, cyan; Gγ, purple; Nb35, grey; ADGRG4-β, slate; ADGRG4-tethered Stachel sequence, hot pink; scFv16, pink; ADGRG2(FL), pale green; ADGRG2 synthetic IP15 peptide, red. b, Cryo-EM map coloured based on local resolution (in Å) for ADGRG2-β–Gs (left panel), ADGRG4-β–Gs (middle panel) and ADGRG2(FL)–IP15–Gs complexes (right panel), respectively. c, Gold-standard Fourier shell correlation (FSC) curves showing an overall resolution at 2.9 Å, 3.3 Å and 3.1 Å for ADGRG2-β–Gs complex (left panel), ADGRG4-β–Gs complex (middle panel) and ADGRG2(FL)–IP15–Gs complex (right panel), respectively. d, Cross-validation of model to cryo-EM density map for ADGRG2-β–Gs complex (left panel), for ADGRG4-β–Gs complex (middle panel) and for ADGRG2(FL)–IP15–Gs complex (right panel), respectively. e, EM density of all seven transmembrane α-helices (TM 1–7), Stachel sequence, α5-helix of Gαs, the helix-8 (H8) and the disulfide bonds formed between the ECL2 and TM3 of ADGRG2-β–Gs complex. Refer to Supplementary Fig. 3, for EM density of ADGRG4-β–Gs complex and ADGRG2(FL)–IP15–Gs complex.
Extended Data Fig. 4 Intramolecular interaction of the Stachel sequences of ADGRG2-β and ADGRG4-β.
a, The Stachel sequence of ADGRG4-β assumed a horizontal “U shape” and laid across the 7TM bundle, which was flanked on one side by TM1, TM6-TM7 and ECL3 and on the other side by TM2-TM3, TM5 and ECL2. The tethered Stachel sequence is shown in pink. b, The Stachel sequence of ADGRG4-β showed a clear division with hydrophilic amino acids (shown in yellow) on the upper rim and hydrophobic residues (shown in pink) on the lower rim and the bottom of the “U shape”. Note that the hydrophobic lower rim and the bottom of the U shape were deeply buried in the 7TM bundle. In contrast, the hydrophilic upper rim of the U shape was solvent exposed. There are no clear EM densities for ECL residues covering the large U shape Stachel sequences, which may enable easy access or dislodgement of the whole U shape Stachel fragments. c, The hydrophobic interactions between the I603 of Stachel sequence (orange stick) and its surrounding residues (green stick) in ADGRG2-β. d, The hydrophobic interactions between the V2726 of Stachel sequence (pink stick) and its surrounding residues (slate stick) in ADGRG4-β. e, f, The five hydrophobic residues F601, I603, L604, L605, and L607 of ADGRG2 (e), and F2724, V2726, L2727, M2728 and L2730 of ADGRG4-β (f) inserted into five hydrophobic pockets in ADGRG2 and ADGRG4, which are highlighted with red dotted circles. g, Detailed interactions within the four hydrophobic pockets of ADGRG4-β that contained the ‘finger’ downward plasma membrane are shown. The tethered Stachel sequences of ADGRG4-β are highlighted in pink. h, Effects of the mutations in the tethered-Stachel-sequence-binding pockets of ADGRG4-β on their constitutive activities were determined by measuring the basal cAMP levels using a GloSensor cAMP assay. The bar graphs were generated according to representative differences between wild-type (WT) ADGRG4-β and its mutants, at a similar expression level of wild-type ADGRG4-β (relative expression level of 8), shown in Supplementary Fig. 6c. Data are normalized and presented as the response percentage for wild-type ADGRG4-β. Values are the mean ± s.e.m. of three independent experiments (n = 3) for wild-type ADGRG4-β and its mutants. Statistical differences between wild-type ADGRG4-β and its mutants were determined by two-sided one-way ANOVA with Tukey’s test. ***P < 0.001; n.s., no significant difference. (P < 0.0001, 0.0001, 0.0005, 0.0002, <0.0001, 0.0002, <0.0001 from top to bottom). i, The whole cell Cl- current elicited by voltage steps between −100 mV and +100 mV in HEK293 cells overexpressing either CFTR and wild-type ADGRG2, or CFTR and ADGRG2-F601ss03A, or CFTR and ADGRG2-L605ss07A, or CFTR and ADGRG2-W771ECL2A, or CFTR and ADGRG2-W8386.53A, in response to stimulation with mechanical forces (cells were treated with shaking at 60 rpm for 2 h according to previous literature). j, Corresponding I-V curves of whole-cell Cl- current as shown in i. Values are the mean ± s.e.m. of six independent experiments (n = 6). k, Average current densities (pA/pF) recorded at 100 mV as shown in j. Values are the mean ± s.e.m. of six independent experiments (n = 6). ***P < 0.001; **P < 0.01; *P < 0.05; HEK293 cells overexpressing CFTR and ADGRG2(FL), or CFTR and ADGRG2 mutants treated with or without shaking were compared. All data were analysed by two-sided one-way ANOVA with Tukey’s test. (P < 0.0001, 0.0396, 0.0036, 0.0011, 0.0226 from left to right). l, Box plots showed the increased current densities (pA/pF) recorded at 100 mV after mechano-forces administration as shown in j. In box plots, whiskers indicate the minimum and maximum values, the bounds of the box are 25th and 75th percentiles of the data, and the centre line indicates the mean. Values are the mean ± s.e.m. of six independent experiments (n = 6). Note that two Stachel sequence mutations (F601ss03A or L605ss07A) and two pocket mutations (W771ECL2A or W8386.53A) significantly decreased the CFTR current in ADGRG2(FL) overexpressed HEK293 cells, supporting that either the functional presence of individual ADGRG2-β alone or the important roles of Stachel sequence mediated interactions in maintaining ADGRG2(FL) activity for CFTR coupling. ***P < 0.001; mechano-forces-stimulated increased current densities in CFTR and ADGRG2 mutants-overexpressed cells were compared with that in CFTR and ADGRG2(FL)-overexpressed cells. All data were analysed by two-sided one-way ANOVA with Tukey’s test. (P < 0.0001, <0.0001, <0.0001, <0.0001 from left to right).
Extended Data Fig. 5 Intramolecular interactions of the Stachel sequence and effects of corresponding mutations.
a, b, Schematic representation of the intramolecular interactions of the Stachel sequence of ADGRG2-β (a) and ADGRG4-β (b). Note that, in ADGRG2-β, the side chains of Y7795.36 and T6211.40 of the upper rim form hydrogen bonds with the main chain of the Stachel sequence, and an intersegment salt bridge was formed by the side chains of D606 and R609. The Stachel sequence in ADGRG4 also anchor with T27471.43. These interactions may help to define the orientation of the peptide and stabilize the overall configuration of the Stachel sequence. Consistent with these observations, elimination of the side chains of T621, Y779 in ADGRG2-β, or T2747 in ADGRG4-β by Ala substitutions significantly impaired the constitutive activity of ADGRG2-β or ADGRG4-β (refer to Supplementary Fig. 5a–f), respectively. The dashed lines with distinct colours represent different interactions (red: charge-charge; green: cation-π; blue: hydrogen bond; orange: polar interaction; black: hydrophobic; pink: π-π). c, Effects of the T599 mutations of ADGRG2-β on intracellular cAMP accumulation at different receptor expression levels. Wild-type ADGRG2-β was used as the control. The intracellular cAMP levels of wild-type ADGRG2-β and its mutants were measured at indicated expression levels as shown in d. Values are the mean ± s.e.m. of three independent experiments for wild-type ADGRG2-β and its mutants (n = 3). Statistical differences between wild-type ADGRG2-β and its mutants were determined by two-sided one-way ANOVA with Tukey’s test, ***P < 0.001; **P < 0.01; n.s., no significant difference. (P = 0.8577, 0.0036, 0.0011, 0.0083, 0.0004, 0.0006 from left to right for T599ss01V; P = 0.3887, 0.0055, 0.0018, 0.0004, <0.0001, 0.0002 from left to right for T599ss01I). d, Similar expression levels of wild-type ADGRG2-β and its T599 mutants are shown by ELISA experiments when HEK293 cells were transiently transfected with indicated amounts of plasmids. Values are the mean ± s.e.m. of three independent experiments for wild-type ADGRG4-β and its mutants (n = 3). Comparisons between the wild-type ADGRG2-β and its mutants were determined by two-sided one-way ANOVA with Tukey’s test. n.s., no significant difference. (P = 0.9839, 0.9763, 0.2183, 0.1553, 0.8098, 0.5447 from left to right for T599ss01V; P = 0.3190, 0.9636, 0.0509, 0.2852, 0.2241, 0.9469 from left to right for T599ss01I). e, RMSD analysis of 200 ns MD simulation of ADGRG2-β, ADGRG2-β-T599V, ADGRG2-β-T599I. Each MD simulation was performed three times (marked with red, dark green and dark blue, respectively). f, The best estimate residue energy contributions of T599V or T599I with 7TM bundle of ADGRG2-β was presented as ΔΔG ((ΔΔG = ΔG Wild-Type - ΔG T599V or T599I)). Values are the mean ± s.e.m. of three independent experiments (n = 3). ***P < 0.001, ADGRG2-β wild-type (WT) and its T599V or T599I mutants were compared. (P < 0.0001, <0.0001 from left to right).
Extended Data Fig. 6 Effects of mutations in the FXφφφXφ motif in ADGRG2-β, ADGRG4-β, ADGRG5-β and ADGRF5-β on their basal activity.
a, The calculated energy contribution of the individual residues within the tethered Stachel sequence region bound in the orthosteric binding pocket of ADGRG4-β. Values are the mean ± s.e.m. of three independent experiments (n = 3). The top five contributing residues are highlighted in red. b, d, e, Effects of mutations in the FXφφφXφ motifs of Stachel sequence in ADGRG4 (GPR112)-β (b), ADGRG5 (GPR114)-β (d) or ADGRF5 (GPR116)-β (e) on their constitutive activities determined by measuring their cAMP levels. Represented differences between wild-type ADGRG4 (GPR112)-β, ADGRG5 (GPR114)-β or ADGRF5 (GPR116)-β and their mutants expressed at a relative expression level of 0.8 (refer to ELISA data for receptor cell surface expression level in Supplementary Fig. 8c–h) are shown as the mean ± s.e.m. of three experiments (n=3). Data are normalized and presented as the response percentage for wild-type ADGRG2-β, ADGRG5-β or ADGRF5-β, respectively. Statistical differences between wild-type and mutant proteins were determined by two-sided one-way ANOVA with Tukey’s test. ***P < 0.001, **P < 0.01; (P < 0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, 0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001 from top to bottom in panel b; P<0.0001, <0.0001, <0.0001, <0.0001, <0.0001, 0.0003, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, 0.0002, <0.0001, <0.0001, <0.0001 from top to bottom in panel d; P = 0.0021, 0.0031, 0.0035, 0.0006, <0.0001, 0.0001, <0.0001, <0.0001, 0.0004, <0.0001, <0.0001, <0.0001 from top to bottom in panel e). FXφφφXφ motif key residues are highlighted in red with blue shades. c, Sequence alignment of Stachel sequence between 33 human adhesion GPCRs. Key residues in the FXφφφXφ motif were labelled by red pentagram. Residues at the positions of FXφφφXφ motif which have large hydrophobic side chains are highlighted in red. Residue numbers following the Stachel sequence (ss) refer to residues related to the position of the GPS. Residues C-terminal to the GPS are numbered starting from 1. f, Cartoon representations of the AlphaFold2 simulated full-length ADGRG4 structure encompassing its GAIN domain. Detailed depictions of the interactions between the buried Stachel sequence of ADGRG4 (pink) in β sheet configuration and its surrounding GAIN domain residues are shown in an enlarged panel on the right. Note that ten hydrophobic residues of the ADGRG4 GAIN domain engaged with the hydrophobic motif of the Stachel sequence of ADGRG4. Through combined MD simulation and chemical labelling, a very recent study6 revealed that the Stachel sequence might be released from the GAIN domain owing to its intrinsic structural flexibility and then be accessible to the 7TM domain. According to this model, the Stachel sequences of ADGRG2 and ADGRG4 might be able to slide away from the GAIN domain and transit to the U-shaped configuration for 7TM domain association in response to changes in different physiological contexts and participate in aGPCR activation. (according to previous publications, the GAIN domain structure of a cleavage-deficient aGPCR should be similar to that of wild-type receptor). g, Effects of mutations in the Stachel sequence-interacting GAIN domain of auto-proteolysis deficient ADGRG4 (GAIN-ADGRG4-β-AA) on their constitutive activities were determined by measuring the basal cAMP levels using a GloSensor cAMP assay. Representative curves for constitutive activities of GAIN-ADGRG4-β-AA and its mutants at indicated expression levels (refer to the ELISA data for the receptor cell surface expression level in Supplementary Fig. 11b) are shown. Values are the mean ± s.e.m. of three independent experiments performed in triplicate (n = 3). Statistical differences between GAIN-ADGRG4-β-AA and its mutants were determined by two-sided one-way ANOVA with Tukey’s test. ***P < 0.001; **P < 0.01; *P < 0.05; n.s., no significant difference (P = 0.2855, 0.0450, 0.0001, 0.0006, 0.0002, <0.0001 from left to right for V2683A curve; P = 0.0535, 0.0112, 0.0092, 0.0345, 0.0027, 0.0003 from left to right; I2660A Curve: P = 0.4548, 0.0135, 0.0004, 0.0257, 0.0026, 0.0002 from left to right for L2671A curve).
Extended Data Fig. 7 Intramolecular interaction between IP15 and ADGRG2(FL).
The therapeutic capacities of peptides derived from Stachel sequence are worth noting. For example, the GPR126 agonist derived from Stachel sequence was able to rescue the phenotype caused by human GPR126 disease associated mutant in a zebrafish model. In addition, Stachel-derived peptides of GPR116/ADGRF5 has therapeutic potential to modulate endogenous alveolar surfactant pools to treat pulmonary diseases associated with surfactant dysfunction. a, Representative dose-response curves for VPM-p15 and IP15 induced cAMP accumulation in HEK293 cells overexpressing ADGRG2-β(ΔS) (ADGRG2-β without Stachel sequence, residues 614–1009) using a GloSensor assay. Values are the mean ± s.e.m. of three independent experiments (n = 3) performed in triplicate. b, EM densities allowed unambiguous assignment of the side chains of the residues in the lower rim of U shape in IP15 (red). The Stachel sequence of ADGRG2 (blue) in ADGRG2-β–Gs complex was aligned with IP15 as a reference. Note that EM density corresponding to upper rim of the U shape was absent compared with the ADGRG2-β–Gs complex structure, and ten out of thirteen residues of the IP15 peptide could be traced according to the EM density. c, The hydrophobic interactions between the I603 of IP15 (red stick) and surrounding residues (pale green sticks) in ADGRG2(FL). d–i, EM densities of the Stachel sequence residues I603 (c), I599 (d), 4-MeF601 (e), L604 (f), L605 (g), L607 (h) (shown in red) and their surrounding residues (shown in green) of ADGRG2(FL) in IP15-ADGRG2(FL)–Gs complex structure. Corresponding residues in the ADGRG2-β–Gs complex was aligned as a reference, Stachel sequence residues are shown in blue, and the surrounding residues are shown in grey. j, RMSD analysis of 4-MeF601 and F601 in 200 ns trajectories. RMSD of 4-MeF601 and its interacting residues during 200 ns MD simulation (upper panel), and RMSD of F601 and its interaction residues during 200 ns MD simulation (lower panel) are shown. k–r, Effects of mutations in IP15 on cAMP accumulation in ADGRG2-β(ΔS) (ADGRG2-β without Stachel sequence, residues 614–1009) overexpressed HEK293 cells. Values are the mean ± s.e.m. of three independent experiments (n = 3) performed in triplicate.
Extended Data Fig. 8 FlAsH BRET-based assays to determine the effects of mutations in IP15 on its association with ADGRG2-β(ΔS) and effects of Stachel-sequence-derived peptidic antagonists on their activities.
a, Representative competition binding curves of ADGRG2-Stachel sequence derived agonist (IP15) or its F601D and F601E mutants (IP15-F601D and IP15-F601E peptides) towards ADGRG2-β(ΔS). Values are the mean ± s.e.m. of three independent experiments (n = 3) performed in triplicate. b, c, Representative dose-dependent inhibition curves of IP15- F601D or IP15- F601E peptide on the constitutive activity of ADGRG2-β (i) or full-length ADGRG2 (j) in terms of cAMP accumulation. Values are the mean ± s.e.m. of three independent experiments (n = 3) performed in triplicate. d, Bar graphs representation of the I/V values of inhibition of whole-cell anion current elicited by voltage steps between −100 mV and +100 mV in HEK293 cells overexpressing with CFTR and mADGRG2, or -CFTR alone. Values are the mean ± s.e.m. of six independent experiments (n = 6). ***P < 0.001, **P < 0.01, n.s., no significant difference. HEK293 cells overexpressing CFTR and ADGRG2, or CFTR-alone treated with or without varying concentrations of IP15-F601D were compared. All data were analysed by two-sided one-way ANOVA with Tukey’s test. (P = 0.1578, 0.0015, 0.0006, 0.4590, 0.4200, 0.8847 from left to right). e, Schematic representation of the FlAsH BRET assay design. Note that the conformational changes in the extracellular domain were reported to be related to the engagement of agonists and antagonists with the 7TM bundle of GPCRs, which correlated to the binding affinities of these ligands. The pcDNA3.1-Nluc-ADGRG2-β plasmid was created by in-fusion the Nluc at the N-terminus of ADGRG2-β, and the FlAsH binding motif (CCPGCC) was incorporated in the designated positions at the extracellular loops (ECLs) of the receptor (S1 and S2 in ECL1; S3 and S4 in ECL2; S5 and S6 in ECL3). Green oval: the position of Nluc; Red pentagram: the binding site of FlAsH. f, Detailed description of the FlAsH motif incorporation sites at the ECLs of ADGRG2 according to 3-dimensional structure shown in i. g, The maximal response of six ADGRG2 FlAsH BRET sensors in response to IP15 or IP15-F601D stimulation. Refer to Supplementary Fig. 13a, b for the FlAsH BRET assay design and corresponding dose-response curves. ND, not detected. Values are the mean ± s.e.m. of three independent experiments (n = 3). h–j, Representative inhibition curves of G4-P15-F2724D, G5-P15-F229D and G6-P15-F843D on the constitutive activity of ADGRG4 (h), ADGRG5 (i) or ADGRG6 (j), respectively. Values are the mean ± s.e.m. of three independent experiments (n = 3) performed in triplicate. k–l, Detailed description of the FlAsH motif incorporation sites at the ECLs of ADGRG4 (k) and ADGRG5 (l). m, n, The maximal response of six ADGRG4 (r) and ADGRG5 (s) FlAsH BRET sensors in response to G4-P15/G5-P15 or G4-P15-F2724D/G5-P15-F229D stimulation. Refer to Supplementary Fig. 13c–f for the FlAsH BRET assay design and corresponding dose-response curves. Values are the mean ± s.e.m. of three independent experiments (n = 3) performed in triplicate. ND, not detected. o, Schematic representation of the agonist and antagonist mode of action on ADGRG2. In response to binding of agonist (IP15) or antagonist (IP15-F601D), the ECL1-S2 and the ECL2-S4 moved close to or away from the N-terminus or ADGRG2, respectively.
Extended Data Fig. 9 The coupling of ADGRG2-β and ADGRG4-β to Gs.
a, Structural superposition of ADGRG2-β–Gs, ADGRG4-β–Gs and ADGRG3–Go (PDB: 7D77) complexes when the TM3 helixes of corresponding receptors were aligned. The orientations of α5 helix of Gs rotated by approximately 9° towards the ADGRG2/ADGRG4 compared to the α5 helix of Go in ADGRG3–Go complex. b, Comparison of the residues between ADGRG2-β, ADGRG4-β and ADGRG3 that contact with their corresponding downstream G proteins (Gs or Go) in cryo-EM structures of ADGRG2-β–Gs, ADGRG4-β–Gs and ADGRG3–Go (PDB: 7D77) complexes, respectively. Contacting residues in ADGRG2-β, ADGRG4-β and ADGRG3 are shown in blue, green and pink dots, respectively, and residues with no interactions are indicated as dashes. Residue positions are labelled with Wootten’s numbers as superscripts on the top of the alignment. Note that ADGRG2 formed more contacts with Gs in the TM2-TM3 region, but the interface showed fewer interactions in the intracellular loops (ICLs) than the interface of GPR97 and Go. c, d, Detailed interactions between Y391 (c) or L393 (d) of the α5 helix of Gαs and ADGRG2-β (left panel) or ADGRG4-β (right panel). ADGRG2-β is shown in green, ADGRG4-β in blue. Hydrogen bonds are depicted as red dashed lines. Note that the side chain of L393 of Gs engaged in hydrophobic packing with V800/V29165.57, L804/L29205.61 and L830/L89456.45 of ADGRG2/ADGRG4, respectively. e, f, Detailed interactions between VFNxY motif in ICL2 of ADGRG2-β (e) or ADGRG4-β (f) and Gs. ADGRG2-β is shown in green, ADGRG4-β in blue, respectively. These interactions included hydrogen bonds, cation-π interactions and extended hydrophobic contacts between the VFNxYICL2 motif of ADGRG2/ADGRG4 and the residues of the αN helix, β1 strand and β3 strand of Gs. Hydrogen bonds are depicted as red dashed lines.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1–11 and Supplementary Figures 1–33.
Rights and permissions
About this article
Cite this article
Xiao, P., Guo, S., Wen, X. et al. Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4. Nature 604, 771–778 (2022). https://doi.org/10.1038/s41586-022-04590-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04590-8
This article is cited by
-
A method for structure determination of GPCRs in various states
Nature Chemical Biology (2024)
-
Constitutive activation mechanism of a class C GPCR
Nature Structural & Molecular Biology (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
The activation mechanism and antibody binding mode for orphan GPR20
Cell Discovery (2023)
-
Mechanism of activation and biased signaling in complement receptor C5aR1
Cell Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.