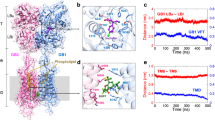
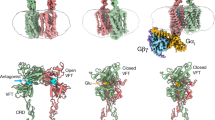
Abstract
Metabotropic γ-aminobutyric acid receptors (GABAB) are involved in the modulation of synaptic responses in the central nervous system and have been implicated in neuropsychological conditions that range from addiction to psychosis1. GABAB belongs to class C of the G-protein-coupled receptors, and its functional entity comprises an obligate heterodimer that is composed of the GB1 and GB2 subunits2. Each subunit possesses an extracellular Venus flytrap domain, which is connected to a canonical seven-transmembrane domain. Here we present four cryo-electron microscopy structures of the human full-length GB1–GB2 heterodimer: one structure of its inactive apo state, two intermediate agonist-bound forms and an active form in which the heterodimer is bound to an agonist and a positive allosteric modulator. The structures reveal substantial differences, which shed light on the complex motions that underlie the unique activation mechanism of GABAB. Our results show that agonist binding leads to the closure of the Venus flytrap domain of GB1, triggering a series of transitions, first rearranging and bringing the two transmembrane domains into close contact along transmembrane helix 6 and ultimately inducing conformational rearrangements in the GB2 transmembrane domain via a lever-like mechanism to initiate downstream signalling. This active state is stabilized by a positive allosteric modulator binding at the transmembrane dimerization interface.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM density maps and corresponding coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and the PDB, respectively, under the following accession codes: EMD-20822 and 6UO8 (SKF97541- and GS39783-bound GABAB), EMD-20823 and 6UO9 (SKF97541-bound GABAB, int-2 state), EMD-20824 and 6UOA (SKF97541-bound GABAB, int-1 state), and EMD-21219 and 6VJM (apo GABAB).
Source data are provided with this paper. Supplementary Figure 1 contains the uncropped gel shown in Extended Data Fig. 1f. All other data are available from the authors on reasonable request.
References
Bowery, N. G. et al. International Union of Pharmacology. XXXIII. Mammalian γ-aminobutyric acidB receptors: structure and function. Pharmacol. Rev. 54, 247–264 (2002).
Comps-Agrar, L. et al. The oligomeric state sets GABAB receptor signalling efficacy. EMBO J. 30, 2336–2349 (2011).
Sieghart, W. Structure, pharmacology, and function of GABAA receptor subtypes. Adv. Pharmacol. 54, 231–263 (2006).
Gassmann, M. & Bettler, B. Regulation of neuronal GABAB receptor functions by subunit composition. Nat. Rev. Neurosci. 13, 380–394 (2012).
Lüscher, C., Jan, L. Y., Stoffel, M., Malenka, R. C. & Nicoll, R. A. G protein-coupled inwardly rectifying K+ channels (GIRKs) mediate postsynaptic but not presynaptic transmitter actions in hippocampal neurons. Neuron 19, 687–695 (1997).
Schuler, V. et al. Epilepsy, hyperalgesia, impaired memory, and loss of pre- and postsynaptic GABAB responses in mice lacking GABAB(1). Neuron 31, 47–58 (2001).
Cousins, M. S., Roberts, D. C. S. & de Wit, H. GABAB receptor agonists for the treatment of drug addiction: a review of recent findings. Drug Alcohol Depend. 65, 209–220 (2002).
Vacher, C. M. & Bettler, B. GABAB receptors as potential therapeutic targets. Curr. Drug Targets CNS Neurol. Disord. 2, 248–259 (2003).
Chang, E. et al. A review of spasticity treatments: pharmacological and interventional approaches. Crit. Rev. Phys. Rehabil. Med. 25, 11–22 (2013).
de Beaurepaire, R. Suppression of alcohol dependence using baclofen: a 2-year observational study of 100 patients. Front. Psychiatry 3, 103 (2012).
Addolorato, G. et al. Baclofen efficacy in reducing alcohol craving and intake: a preliminary double-blind randomized controlled study. Alcohol Alcohol. 37, 504–508 (2002).
Lapin, I. Phenibut (β-phenyl-GABA): a tranquilizer and nootropic drug. CNS Drug Rev. 7, 471–481 (2001).
Dalmau, J. & Graus, F. Antibody-mediated encephalitis. N. Engl. J. Med. 378, 840–851 (2018).
Hamdan, F. F. et al. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am. J. Hum. Genet. 101, 664–685 (2017).
Vuillaume, M.-L. et al. A novel mutation in the transmembrane 6 domain of GABBR2 leads to a Rett-like phenotype. Ann. Neurol. 83, 437–439 (2018)
Yoo, Y. et al. GABBR2 mutations determine phenotype in Rett syndrome and epileptic encephalopathy. Ann. Neurol. 82, 466–478 (2017).
Kniazeff, J., Prézeau, L., Rondard, P., Pin, J.-P. & Goudet, C. Dimers and beyond: the functional puzzles of class C GPCRs. Pharmacol. Ther. 130, 9–25 (2011).
Stewart, G. D., Comps-Agrar, L., Nørskov-Lauritsen, L. B., Pin, J. P. & Kniazeff, J. Allosteric interactions between GABAB1 subunits control orthosteric binding sites occupancy within GABAB oligomers. Neuropharmacology 136, 92–101 (2018).
White, J. H. et al. Heterodimerization is required for the formation of a functional GABAB receptor. Nature 396, 679–682 (1998).
Margeta-Mitrovic, M., Jan, Y. N. & Jan, L. Y. A trafficking checkpoint controls GABAB receptor heterodimerization. Neuron 27, 97–106 (2000).
Galvez, T. et al. Allosteric interactions between GB1 and GB2 subunits are required for optimal GABAB receptor function. EMBO J. 20, 2152–2159 (2001).
Robbins, M. J. et al. GABAB2 is essential for G-protein coupling of the GABAB receptor heterodimer. J. Neurosci. 21, 8043–8052 (2001).
Geng, Y., Bush, M., Mosyak, L., Wang, F. & Fan, Q. R. Structural mechanism of ligand activation in human GABAB receptor. Nature 504, 254–259 (2013).
Pin, J.-P. & Bettler, B. Organization and functions of mGlu and GABAB receptor complexes. Nature 540, 60–68 (2016).
Howson, W., Mistry, J., Broekman, M. & Hills, J. M. Biological activity of 3-aminopropyl (methyl) phosphinic acid, a potent and selective GABAB agonist with CNS activity. Bioorg. Med. Chem. Lett. 3, 515–518 (1993).
Frankowska, M., Filip, M. & Przegaliński, E. Effects of GABAB receptor ligands in animal tests of depression and anxiety. Pharmacol. Rep. 59, 645–655 (2007).
Cryan, J. F. et al. Behavioral characterization of the novel GABAB receptor-positive modulator GS39783 (N,N′-dicyclopentyl-2-methylsulfanyl-5-nitro-pyrimidine-4,6-diamine): anxiolytic-like activity without side effects associated with baclofen or benzodiazepines. J. Pharmacol. Exp. Ther. 310, 952–963 (2004).
Koehl, A. et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019).
Isberg, V. et al. Generic GPCR residue numbers – aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 36, 22–31 (2015).
Xue, L. et al. Rearrangement of the transmembrane domain interfaces associated with the activation of a GPCR hetero-oligomer. Nat. Commun. 10, 2765 (2019).
Duthey, B. et al. A single subunit (GB2) is required for G-protein activation by the heterodimeric GABAB receptor. J. Biol. Chem. 277, 3236–3241 (2002).
Urwyler, S. et al. N,N′-Dicyclopentyl-2-methylsulfanyl-5-nitro-pyrimidine-4,6-diamine (GS39783) and structurally related compounds: novel allosteric enhancers of γ-aminobutyric acidB receptor function. J. Pharmacol. Exp. Ther. 307, 322–330 (2003).
Monnier, C. et al. Trans-activation between 7TM domains: implication in heterodimeric GABAB receptor activation. EMBO J. 30, 32–42 (2011).
Kniazeff, J. et al. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat. Struct. Mol. Biol. 11, 706–713 (2004).
Schorb, M., Haberbosch, I., Hagen, W. J. H., Schwab, Y. & Mastronarde, D. N. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Huang, J. & MacKerell, A. D. Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Abagyan, R., Totrov, M. & Kuznetsov, D. ICM: a new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506 (1994).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Kim, S. et al. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 38, 1879–1886 (2017).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Bakan, A., Meireles, L. M. & Bahar, I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27, 1575–1577 (2011).
Gur, M., Zomot, E. & Bahar, I. Global motions exhibited by proteins in micro- to milliseconds simulations concur with anisotropic network model predictions. J. Chem. Phys. 139, 121912 (2013).
Krissinel, E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43, W314–W319 (2015).
Acknowledgements
We thank K. Villers, C. Hanson, F. Nasertorabi, J. Velasquez and M. Barekatain for technical support; S. Roux for the help with collecting functional data; E. Montabana for microscope support; Y.-T. Li and N. Patel for support with computing resources; and Y. Kadyshevskaya for help with illustrations. This research was supported by the National Institutes of Health (NIH) grant R35 GM127086 (V.C.), by the Department of Energy, Laboratory Directed Research and Development program at SLAC National Accelerator Laboratory, under contract DE-AC02-76SF00515, and by the Fondation pour la Recherche Médicale (DEQ20170336747, to J.-P.P.). We acknowledge the SLAC-Stanford Cryo-EM center (SLAC National Accelerator Laboratory) for microscope time. Functional experiments were performed at the ARPEGE (Pharmacology Screening-Interactome) facility, Institut de Génomique Fonctionnelle. We acknowledge Google Cloud Platform credits and USC High Performance Computing Center for computer resources used in molecular dynamics simulations.
Author information
Authors and Affiliations
Contributions
H.S. expressed and purified GABAB heterodimers for cryo-EM data collection. A.I. helped with the development of protocols for receptor expression and purification. C.G. performed cryo-EM sample preparation, data collection and image processing. L.X. performed ligand-binding and functional experiments. G.W.H. helped with the structure refinement and quality control. J.H.L, and V.K. performed molecular dynamics and ANM modelling. P.R., J.-P.P., J.H.L. and V.K. analysed data. H.S., C.G. and V.C. conceived the project, analysed data, and wrote the manuscript with contributions from all authors. V.C. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Expression, characterization and purification of GABAB.
a, b, Cell surface expression of Halo–GB1 (a) and SNAP–GB2 (b), transfected alone or cotransfected with the second subunit, measured by the fluorescence emission of the Lumi4-Tb bound to the Halo (a) or SNAP tag (b). Values are normalized by the wild-type GB1 cotransfected with wild-type GB2 (purple bar) and shown as mean ± s.d. of three biologically independent experiments. GABAB constructs for cryo-EM are expressed and function similarly to the wild-type receptor. c, d, Positive allosteric effect of GS39783 (5 μM) on IP1 accumulation in cells expressing wild-type or cryo-EM constructs of GABAB heterodimers and activated either by GABA (c) or SKF97541 (d). Data are normalized by the wild-type response in absence of GS39783 (control) and shown as mean ± s.e.m. of three biologically independent experiments (4 for wild type). e, Representative SEC profile of apo GABAB in digitonin micelles. Dimeric fractions were pooled, supplemented with ligand and concentrated for cryo-EM imaging. f, Coomassie stained SDS–PAGE profile shows two distinct bands for GB1 (86 kDa) and GB2 (88 kDa). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Cryo-EM data processing for SKF97541-bound GABAB.
a, Single-particle cryo-EM data processing scheme using RELION and cryoSPARC. b, Representative micrograph showing picked particles. c, Representative 2D class averages. d, Angular distribution of particles included in the final cryo-EM reconstruction for the int-1 state. e, The same as d but for the int-2 state. f, g, Density maps for int-1 (f) and int-2 (g) states coloured by local resolution, showing higher resolution at the VFT interface.
Extended Data Fig. 3 Cryo-EM map quality.
a–d, Representative densities and fitted atomic models are shown for GABAB in the inactive apo state (a), active agonist- and PAM-bound state (b), and two intermediate int-1 (c) and int-2 states (d).
Extended Data Fig. 4 Cryo-EM data processing for SKF97541- and GS39783-bound GABAB.
a, Single-particle cryo-EM data processing scheme using cryoSPARC. b, Micrograph showing picked particles. c, Representative 2D class averages show distinct secondary structure features. d, Particle angular distribution of the final cryo-EM reconstruction. e, Structural model shown in cartoon representation and coloured by the B-factors. f, Density map coloured by local resolution, showing higher resolution at the interface between GB1 and GB2. g, The heterodimer interface includes both LB1 and LB2, as well as TM6, with a total interface area of 1,930 Å2. Residues involved in dimerization contacts are coloured in purple.
Extended Data Fig. 5 Cryo-EM data processing for apo GABAB.
a, Single-particle cryo-EM data processing scheme using cryoSPARC. b, Micrograph showing picked particles. c, Representative 2D class averages show distinct secondary structure features. d, Particle angular distribution of the final cryo-EM reconstruction. e, Structural model shown in cartoon representation and coloured by the B-factors. f, Density map coloured by local resolution, showing higher resolution at the interface of GB1 and GB2. g, Gold-standard FSC curves from cryoSPARC. h, Map-to-model FSC curve between each refined model and the corresponding sharpened electron potential maps. i, GB1 and GB2 interact through LB1s and intracellular tips of TM3 and TM5, with a total interface area of 963 Å2. Residues involved in dimerization contacts are coloured in purple.
Extended Data Fig. 6 Molecular dynamics simulations of inactive (apo) and active (agonist- and PAM-bound) states.
a, VFT fluctuations in the inactive state are illustrated by comparing molecular dynamics snapshots at 500 ns for all 5 trajectories (cartoon, purple for α-helices and yellow for β-strands) against the cryo-EM structure (grey surface). Cyan spheres indicate nitrogen of hydrophilic heads of the POPC lipid bilayer. Asterisk corresponds to trajectory 1. b, Traces of Cα r.m.s.d. for different domains in trajectory 1. The structures in the trajectory were aligned by TMDs against the cryo-EM structure. c, Distribution of distances for ionic interactions between TM3 and TM5 at the intracellular side of the apo GABAB, as observed in molecular dynamics simulations. d, e, Membrane-coupled ANM analysis of the inactive (d)- and active (e)-state cryo-EM structures. Eigenvectors of the slowest mode are drawn on every eighth Cα as magenta arrows. In the inset heat map, overall dynamics of the protein as observed in molecular dynamics simulations are compared against that predicted from ANM analysis on electron microscopy structures; correlations between eigenvectors of the ten slowest ANM modes and the principal components of Cα motions in molecular dynamics simulations are calculated, showing concurrence between the two. f, Distance plots as observed in agonist- and PAM-bound trajectories for interactions between SKF97541 (SKF) and various contact residues in the GB1 pocket (left column), interactions between GS39783 (PAM) and its contact residues at the TMD interface (right column, rows 1–3) and interactions between GB1 and GB2 residues (right column, rows 4 and 5). g, h, Traces of Cα r.m.s.d. on the overall structure for individual molecular dynamics trajectories (green) and plots of root mean square fluctuations (r.m.s.f.) per residue in GB1 (blue) and GB2 (yellow) subunits as observed in molecular dynamics simulations for inactive (g) and active (h) states. i, j, Stacked snapshots of GS39783 (i) and SKF97541 (j) binding site for one of the trajectories, taken every 50 ns and spanning 500 ns. In traces of Cα r.m.s.d. (b, g, h), data (solid line) are presented as an average within a sliding window of 500 ps; shading refers to 95% confidence interval (n = 500). In plots of r.m.s.f. (g, h), data (solid line) are shown as an average of 5 independent trajectories at each residue position; shading refers to 95% confidence interval (n = 5).
Extended Data Fig. 7 Comparisons with previous crystal structures and additional details of activation-related transitions in GABAB.
a, Comparison of the VFTs between the apo cryo-EM structure (teal) and the apo crystal structure (PDB 4MQE, grey). b, Comparison of the VFTs between the active agonist- and PAM-bound cryo-EM structure (orange) and the GABA-bound crystal structure (PDB 4MS3, grey). c, Comparison between GB1 and GB2 TMDs in the inactive apo state demonstrates no substantial difference between the subunits. d, Comparison between GB1 and GB2 TMDs in the active agonist- and PAM-bound state illustrates the differences between the subunits upon activation. e, The TMD of GB1 remains unchanged between the int-2 and the active agonist- and PAM-bound states. f, Shifts of TM3 and TM5 at the intracellular side of the TMD of GB2 between the int-2 and active states open up a cleft for the engagement of a G protein.
Extended Data Fig. 8 Effects of a PAM on the orthosteric ligand binding in the site-1 and -2 mutants of GABAB.
a, Displacement of the CGP54626-DY647 by GABA in intact cells expressing the wild-type GABAB receptor in the absence (blue) or presence of 5 μM (red), 10 μM (orange) or 20 μM (sand) GS39783. b, Cell-surface expression of site-1 and -2 mutants, measured by cotransfecting Halo–GB1 with GB2 and recording fluorescence emission of Halo–Lumi4-Tb. Data are normalized by the wild-type expression and shown as mean ± s.d. The numbers of biologically independent experiments are shown in parentheses. c, pKi values for GABA were determined from displacement of CGP54626-DY647 binding in intact cells expressing the indicated subunit combinations in the absence or presence of 1 μM or 5 μM GS39783. Values are mean ± s.e.m. of 4 (10 for wild type) biologically independent experiments. Data are analysed using one-way ANOVA with Dunnett’s multiple comparison test to determine significance (compared with no GS39783 for the same combined subunits), with ****P ≤ 0.0001, ***P ≤ 0.001, *P ≤ 0.01 and NS (not significant) P > 0.05. For 1 μM (5 μM) GS39783, P = 0.0030 (0.0045) for GB2(R556A), 0.0205 (0.0157) for GB2(H647A), 0.0084 (0.0009) for GB2(L657A), 0.0436 (0.0003) for GB2(F711A), 0.0013 (0.0053) for GB2(R714A), 0.0373 (0.0056) for GB2(Q720A), and 0.0735 (0.0019) for GB2(SFR-AAA). GB2(SFR-AAA) refers to a triple mutant (S710A/F711A/R714A) of GB2. d, e, Displacement of the CGP54626-DY647 by GABA in intact cells expressing the indicated mutants in site 1 (d) and site 2 (e), in the absence or presence of the indicated concentrations of GS39783 as in a. Data are normalized by the signal in absence of GS39783 and shown as mean ± s.e.m. of three biologically independent experiments.
Extended Data Fig. 9 The effects of a PAM on signalling of the site-1 and -2 mutants of GABAB.
a, IP1 production induced by 10 μM GS39783 in intact cells expressing the indicated site-1 GB2 mutants co-expressed with the wild-type GB1. Data are normalized by the GABA response and shown as mean ± s.d. of 7 (10 for wild type) biologically independent experiments. Data are analysed using one-way ANOVA with Dunnett’s multiple comparison test to determine significance (compared with wild type), with P = 0.9998 for GB2(R556A), 0.9999 for GB2(H647A), 0.9907 for GB2(L657A), 0.9995 for GB1(F711A), 0.9997 for GB2(R714A), 0.9996 for GB2(Q720A), and 0.2807 for GB2(SFR-AAA). b–d, IP1 production induced by GABA (blue) or GS39783 (red) in intact cells expressing the wild-type receptor (b) or the indicated mutant of the site 1 (c) or site 2 (d). Data are normalized by the GABA response and shown as mean ± s.e.m. of four biologically independent experiments.
Supplementary information
Supplementary Figure 1
This file contains the uncropped gel used in Extended Data Fig. 1f.
Video 1
: Conformational transitions related to GABAB activation Overview of the modeled activation pathway as illustrated transitions between the four obtained models of GABAB heterodimers. Morphs were calculated between inactive apo, agonist-bound int-1 and int-2, to the agonist/PAM-bound structure, with a sequential alignment of the entire receptor.
Video 2
: Details of SKF97541 binding Rocking video (90°) around the electron potential map of the agonist SKF97541 in the binding pocket of the agonist/PAM-bound GABAB structure, depicted in Fig. 3c.
Video 3
: Details of GS39783 binding Rocking video (60°) around the electron potential map of PAM GS39783 in the binding pocket at the dimer interface of the agonist/PAM-bound GABAB structure, depicted in Fig. 3d.
Rights and permissions
About this article
Cite this article
Shaye, H., Ishchenko, A., Lam, J.H. et al. Structural basis of the activation of a metabotropic GABA receptor. Nature 584, 298–303 (2020). https://doi.org/10.1038/s41586-020-2408-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2408-4
This article is cited by
-
Specific pharmacological and Gi/o protein responses of some native GPCRs in neurons
Nature Communications (2024)
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Filamin A organizes γ‑aminobutyric acid type B receptors at the plasma membrane
Nature Communications (2023)
-
G protein-coupled receptors in neurodegenerative diseases and psychiatric disorders
Signal Transduction and Targeted Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.