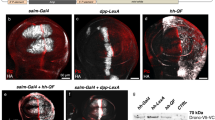
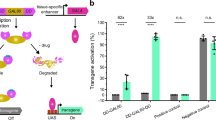
Abstract
Tissue-specific gene expression using the upstream activating sequence (UAS)–GAL4 binary system has facilitated genetic dissection of many biological processes in Drosophila melanogaster. Refining GAL4 expression patterns or independently manipulating multiple cell populations using additional binary systems are common experimental goals. To simplify these processes, we developed a convertible genetic platform, the integrase swappable in vivo targeting element (InSITE) system. This approach allows GAL4 to be replaced with any other sequence, placing different genetic effectors under the control of the same regulatory elements. Using InSITE, GAL4 can be replaced with LexA or QF, allowing an expression pattern to be repurposed. GAL4 can also be replaced with GAL80 or split-GAL4 hemi-drivers, allowing intersectional approaches to refine expression patterns. The exchanges occur through efficient in vivo manipulations, making it possible to generate many swaps in parallel. This system is modular, allowing future genetic tools to be easily incorporated into the existing framework.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brand, A.H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
O'Kane, C.J. & Gehring, W.J. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 84, 9123–9127 (1987).
Ejsmont, R.K., Sarov, M., Winkler, S., Lipinski, K.A. & Tomancak, P. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods 6, 435–437 (2009).
Pfeiffer, B.D. et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715–9720 (2008).
Venken, K.J. et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434 (2009).
Luo, L., Callaway, E.M. & Svoboda, K. Genetic dissection of neural circuits. Neuron 57, 634–660 (2008).
Korzh, V. Transposons as tools for enhancer trap screens in vertebrates. Genome Biol. 8 (Suppl. 1), S8 (2007).
Stanford, W.L., Cohn, J.B. & Cordes, S.P. Gene-trap mutagenesis: past, present and beyond. Nat. Rev. Genet. 2, 756–768 (2001).
Bellen, H.J. Ten years of enhancer detection: lessons from the fly. Plant Cell 11, 2271–2281 (1999).
Lai, S.L. & Lee, T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 9, 703–709 (2006).
Potter, C.J., Tasic, B., Russler, E.V., Liang, L. & Luo, L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536–548 (2010).
Suster, M.L., Seugnet, L., Bate, M. & Sokolowski, M.B. Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis 39, 240–245 (2004).
Bohm, R.A. et al. A genetic mosaic approach for neural circuit mapping in Drosophila. Proc. Natl. Acad. Sci. USA 107, 16378–16383 (2010).
Stockinger, P., Kvitsiani, D., Rotkopf, S., Tirian, L. & Dickson, B.J. Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807 (2005).
Gordon, M.D. & Scott, K. Motor control in a Drosophila taste circuit. Neuron 61, 373–384 (2009).
Luan, H., Peabody, N.C., Vinson, C.R. & White, B.H. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436 (2006).
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999).
Bateman, J.R., Lee, A.M. & Wu, C.T. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173, 769–777 (2006).
Bischof, J., Maeda, R.K., Hediger, M., Karch, F. & Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317 (2007).
Golic, M.M., Rong, Y.S., Petersen, R.B., Lindquist, S.L. & Golic, K.G. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 25, 3665–3671 (1997).
Horn, C. & Handler, A.M. Site-specific genomic targeting in Drosophila. Proc. Natl. Acad. Sci. USA 102, 12483–12488 (2005).
Oberstein, A., Pare, A., Kaplan, L. & Small, S. Site-specific transgenesis by Cre-mediated recombination in Drosophila. Nat. Methods 2, 583–585 (2005).
Schlake, T. & Bode, J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33, 12746–12751 (1994).
Yagi, R., Mayer, F. & Basler, K. Refined LexA transactivators and their use in combination with the Drosophila Gal4 system. Proc. Natl. Acad. Sci. USA 107, 16166–16171 (2010).
Groth, A.C., Fish, M., Nusse, R. & Calos, M.P. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 (2004).
Thorpe, H.M. & Smith, M.C. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. USA 95, 5505–5510 (1998).
Woltjen, K. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 (2009).
Horn, C., Offen, N., Nystedt, S., Hacker, U. & Wimmer, E.A. piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics 163, 647–661 (2003).
Rad, R. et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 330, 1104–1107 (2010).
Li, X. et al. piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol. Biol. 14, 17–30 (2005).
Thibault, S.T. et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283–287 (2004).
Siegal, M.L. & Hartl, D.L. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144, 715–726 (1996).
Maggert, K.A., Gong, W.J. & Golic, K.G. Methods for homologous recombination in Drosophila. Methods Mol. Biol. 420, 155–174 (2008).
Gao, S. et al. The neural substrate of spectral preference in Drosophila. Neuron 60, 328–342 (2008).
Newsome, T.P., Asling, B. & Dickson, B.J. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851–860 (2000).
Venken, K.J., He, Y., Hoskins, R.A. & Bellen, H.J. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006).
Malla, S., Dafhnis-Calas, F., Brookfield, J.F., Smith, M.C. & Brown, W.R. Rearranging the centromere of the human Y chromosome with phiC31 integrase. Nucleic Acids Res. 33, 6101–6113 (2005).
Pfeiffer, B.D. et al. Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010).
Awasaki, T., Lai, S.L., Ito, K. & Lee, T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J. Neurosci. 28, 13742–13753 (2008).
Gohl, D., Muller, M., Pirrotta, V., Affolter, M. & Schedl, P. Enhancer blocking and transvection at the Drosophila apterous locus. Genetics 178, 127–143 (2008).
Schuldiner, O. et al. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell 14, 227–238 (2008).
Hagstrom, K., Muller, M. & Schedl, P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10, 3202–3215 (1996).
Rubin, G.M. & Spradling, A.C. Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–353 (1982).
Potter, C.J. & Luo, L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS ONE 5, e10168 (2010).
Ochman, H., Gerber, A.S. & Hartl, D.L. Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623 (1988).
Tweedie, S. et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37, D555–D559 (2009).
Wagh, D.A. et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844 (2006).
Acknowledgements
We thank members of the Clandinin laboratory for helpful advice; M. Müller (University of Basel), M. Wernet (Stanford University), T. Schwabe (Stanford University), G. Dietzl (Stanford University), J. Bateman (Bowdoin College), L. Luo (Stanford University), C.-H. Lee (US National Institutes of Health) and P. Schedl (Princeton University) for reagents; S. Burns for assistance with experiments; and A. Parks at the Bloomington Drosophila Stock Center. M. Klovstad, L. Luo and T. Schwabe provided valuable comments on the manuscript. M. Spletter helped score antennal lobes. This work was funded by a National Institutes of Health Director's Pioneer Award DP1 OD003530 (T.R.C.) and by National Institutes of Health R01 EY015231 (T.R.C.). D.M.G. and M.A.S. were supported by Stanford Dean's Postdoctoral fellowships.
Author information
Authors and Affiliations
Contributions
D.M.G. and T.R.C. designed the experiments; D.M.G., M.A.S., X.J.G., F.J.L., C.-C.L., C.J.P. and T.R.C. performed the experiments; S.B. provided critical reagents; and D.M.G. and T.R.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1–3 (PDF 5749 kb)
Rights and permissions
About this article
Cite this article
Gohl, D., Silies, M., Gao, X. et al. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods 8, 231–237 (2011). https://doi.org/10.1038/nmeth.1561
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1561
This article is cited by
-
Dopaminergic systems create reward seeking despite adverse consequences
Nature (2023)
-
Copy number changes in co-expressed odorant receptor genes enable selection for sensory differences in drosophilid species
Nature Ecology & Evolution (2022)
-
SPARC enables genetic manipulation of precise proportions of cells
Nature Neuroscience (2020)
-
Olfactory receptor and circuit evolution promote host specialization
Nature (2020)
-
Wrapping glia regulates neuronal signaling speed and precision in the peripheral nervous system of Drosophila
Nature Communications (2020)