Abstract
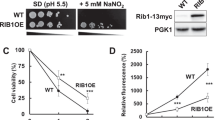
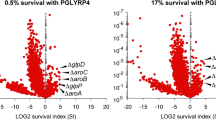
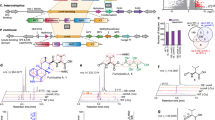
Nitric oxide (NO) is a toxic reactive nitrogen species that induces microbial adaption mechanisms. Screening a genomic DNA library identified a new gene, ntpA, that conferred growth tolerance upon Aspergillus nidulans against exogenous NO. The gene encoded a cysteine-rich 23-amino-acid peptide that reacted with NO and S-nitrosoglutathione to generate an S-nitrosated peptide. Disrupting ntpA increased amounts of cellular S-nitrosothiol and NO susceptibility. Thioredoxin and its reductase denitrosated the S-nitrosated peptide, decreased cellular S-nitrosothiol and conferred tolerance against NO, indicating peptide-mediated catalytic NO removal. The peptide binds copper(I) in vitro but is dispensable for metal tolerance in vivo. NO but not metal ions induced production of the peptide and ntpA transcripts. We discovered that the thionein family of peptides has NO-related functions and propose that the new peptide be named NO-inducible nitrosothionein (iNT). The ubiquitous distribution of iNT-like polypeptides constitutes a potent NO-detoxifying mechanism that is conserved among various organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Stamler, J.S. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell 78, 931–936 (1994).
Goretski, J., Zafiriou, O.C. & Hollocher, T.C. Steady-state nitric oxide concentrations during denitrification. J. Biol. Chem. 265, 11535–11538 (1990).
Hausladen, A. & Stamler, J.S. Nitrosative stress. Methods Enzymol. 300, 389–395 (1999).
Forrester, M.T. & Foster, M.W. Protection from nitrosative stress: a central role for microbial flavohemoglobin. Free Radic. Biol. Med. 52, 1620–1633 (2012).
Gardner, A.M., Helmick, R.A. & Gardner, P.R. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 277, 8172–8177 (2002).
Shoun, H. & Tanimoto, T. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266, 11078–11082 (1991).
St. John, G. et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98, 9901–9906 (2001).
De Groote, M.A., Testerman, T., Xu, Y., Stauffer, G. & Fang, F.C. Homocysteine antagonism of nitric oxide–related cytostasis in Salmonella typhimurium. Science 272, 414–417 (1996).
Flatley, J. et al. Transcriptional responses of Escherichia coli to S-nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J. Biol. Chem. 280, 10065–10072 (2005).
Hromatka, B.S., Noble, S.M. & Johnson, A.D. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16, 4814–4826 (2005).
Nittler, M.P., Hocking-Murray, D., Foo, C.K. & Sil, A. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol. Biol. Cell 16, 4792–4813 (2005).
Sarver, A. & DeRisi, J. Fzf1p regulates an inducible response to nitrosative stress in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 4781–4791 (2005).
Hess, D.T., Matsumoto, A., Kim, S.O., Marshall, H.E. & Stamler, J.S. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 (2005).
Clancy, R.M., Levartovsky, D., Leszczynska-Piziak, J., Yegudin, J. & Abramson, S.B. Nitric oxide reacts with intracellular glutathione and activates the hexose monophosphate shunt in human neutrophils: evidence for S-nitrosoglutathione as a bioactive intermediary. Proc. Natl. Acad. Sci. USA 91, 3680–3684 (1994).
Liu, L. et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 (2001).
Benhar, M., Forrester, M.T. & Stamler, J.S. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732 (2009).
Foster, M.W., Liu, L., Zeng, M., Hess, D.T. & Stamler, J.S. A genetic analysis of nitrosative stress. Biochemistry 48, 792–799 (2009).
Wei, W. et al. S-Nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci. Transl. Med. 2, 19ra13 (2010).
Takasaki, K. et al. Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. Role of acetyl CoA synthetase in anaerobic ATP synthesis. J. Biol. Chem. 279, 12414–12420 (2004).
Shimizu, M., Fujii, T., Masuo, S., Fujita, K. & Takaya, N. Proteomic analysis of Aspergillus nidulans cultured under hypoxic conditions. Proteomics 9, 7–19 (2009).
Zhou, S. et al. Functional analysis and subcellular location of two flavohemoglobins from Aspergillus oryzae. Fungal Genet. Biol. 48, 200–207 (2011).
de Jesús-Berríos, M. et al. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13, 1963–1968 (2003).
Schinko, T. et al. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol. Microbiol. 78, 720–738 (2010).
Zhou, S. et al. Heme-biosynthetic porphobilinogen deaminase protects Aspergillus nidulans from nitrosative stress. Appl. Environ. Microbiol. 78, 103–109 (2012).
Wortman, J.R. et al. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet. Biol. 46 (suppl. 1): S2–S13 (2009).
Capdevila, M. & Atrian, S. Metallothionein protein evolution: a miniassay. J. Biol. Inorg. Chem. 16, 977–989 (2011).
de Oliveira, M.G., Shishido, S.M., Seabra, A.B. & Morgon, N.H. Thermal stability of primary S-nitrosothiols: roles of autocatalysis and structural effects on the rate of nitric oxide release. J. Phys. Chem. A 106, 8963–8970 (2002).
Jaffrey, S.R. & Snyder, S.H. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001, pl1 (2001).
Gow, A., Doctor, A., Mannick, J. & Gaston, B. S-Nitrosothiol measurements in biological systems. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 851, 140–151 (2007).
Romero, J.M. & Bizzozero, O.A. Intracellular glutathione mediates the denitrosylation of protein nitrosothiols in the rat spinal cord. J. Neurosci. Res. 87, 701–709 (2009).
Stoyanovsky, D.A. et al. Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J. Am. Chem. Soc. 127, 15815–15823 (2005).
Thön, M., Al-Abdallah, Q., Hortschansky, P. & Brakhage, A.A. The thioredoxin system of the filamentous fungus Aspergillus nidulans: impact on development and oxidative stress response. J. Biol. Chem. 282, 27259–27269 (2007).
Sengupta, R. et al. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry 46, 8472–8483 (2007).
Benhar, M., Forrester, M.T., Hess, D.T. & Stamler, J.S. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320, 1050–1054 (2008).
Nikitovic, D. & Holmgren, A. S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 271, 19180–19185 (1996).
Thön, M. et al. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 38, 1098–1113 (2010).
Gold, B. et al. Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat. Chem. Biol. 4, 609–616 (2008).
Schwarz, M.A. et al. Metallothionein protects against the cytotoxic and DNA-damaging effects of nitric oxide. Proc. Natl. Acad. Sci. USA 92, 4452–4456 (1995).
Tucker, S.L. et al. A fungal metallothionein is required for pathogenicity of Magnaporthe grisea. Plant Cell 16, 1575–1588 (2004).
Münger, K., Germann, U.A. & Lerch, K. Isolation and structural organization of the Neurosporra crassa copper metallothionein gene. EMBO J. 4, 2665–2668 (1985).
Vašák, M. & Meloni, G. Chemistry and biology of mammalian metallothioneins. J. Biol. Inorg. Chem. 16, 1067–1078 (2011).
Liu, S.X. et al. Reconstitution of apo-superoxide dismutase by nitric oxide-induced copper transfer from metallothioneins. Chem. Res. Toxicol. 13, 922–931 (2000).
Cai, L., Koropatnick, J. & Cherian, M.G. Metallothionein protects DNA from copper-induced but not iron-induced cleavage in vitro. Chem. Biol. Interact. 96, 143–155 (1995).
Kröncke, K.D. Nitrosative stress and transcription. Biol. Chem. 384, 1365–1377 (2003).
Lushchak, O.V., Inoue, Y. & Lushchak, V.I. Regulatory protein Yap1 is involved in response of yeast Saccharomyces cerevisiae to nitrosative stress. Biochemistry (Mosc). 75, 629–664 (2010).
Asano, Y., Hagiwara, D., Yamashino, T. & Mizuno, T. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71, 1800–1803 (2007).
Datta, P.K. & Lianos, E.A. Nitric oxide induces metallothionein-1 gene expression in mesangial cells. Transl. Res. 148, 180–187 (2006).
Liu, L. et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116, 617–628 (2004).
Acknowledgements
We thank H. Shoun and S. Fushinobu for help with fluorometry and S. Osmani for his valuable suggestions in fungal transformation. We thank N. Foster for helpful discussion and critical reading of the manuscript. This study was supported by Grants-in-Aid for Scientific Research, Japan Society for the Promotion of Science (23-01090 to S.Z. and 21380055 to N.T.).
Author information
Authors and Affiliations
Contributions
N.T. and S.Z. planned the studies and prepared the manuscript. S.Z., T.N. and Y.K. designed and performed experiments. S.M., T.N., M.S., T.F., Y.D. and Y.K. performed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1 and 2 and Supplementary Figures 1–9 (PDF 1060 kb)
Rights and permissions
About this article
Cite this article
Zhou, S., Narukami, T., Masuo, S. et al. NO-inducible nitrosothionein mediates NO removal in tandem with thioredoxin. Nat Chem Biol 9, 657–663 (2013). https://doi.org/10.1038/nchembio.1316
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1316
This article is cited by
-
Mycothiol maintains the homeostasis and signalling of nitric oxide in Streptomyces coelicolor A3(2) M145
BMC Microbiology (2023)
-
Interplay of two transcription factors for recruitment of the chromatin remodeling complex modulates fungal nitrosative stress response
Nature Communications (2021)
-
Thiol-based redox-active proteins as cardioprotective therapeutic agents in cardiovascular diseases
Basic Research in Cardiology (2021)
-
A Novel Mechanism for Nitrosative Stress Tolerance Dependent on GTP Cyclohydrolase II Activity Involved in Riboflavin Synthesis of Yeast
Scientific Reports (2020)
-
Nitric oxide in fungi: is there NO light at the end of the tunnel?
Current Genetics (2016)