Abstract
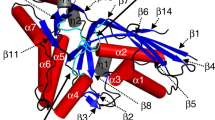
The crystal structure of 5'-nucleotidase (5'-NT) from E. coli, also known as UDP-sugar hydrolase, has been determined at 1.7 Å resolution. Two zinc ions are present in the active site, which is located in a cleft between two domains. The dimetal center and a catalytic Asp-His dyad are the main players in the catalytic mechanism. Structure-based sequence comparisons show that the structure also provides a model for animal 5'-NTs, which together with other ectonucleotidases terminate the action of nucleotides as extracellular signaling substances in the nervous system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Zimmermann, H. Biochem. J. 285, 345–365 (1992).
Zimmermann, H. Prog. Neurobiol. 49, 589–618 (1996).
Kennedy, C., Todorov, L.D., Mihaylova-Todorova, S. & Sneddon, P. Trends Pharmacol. Sci. 18, 263–266 (1997).
Pearson, J.D., Carleton, J.S. & Gordon, J.L. Biochem. J. 190, 421– 429 (1987).
Zimmermann, H., Dowdall, M.J. & Lane, D.A. Neuroscience 4, 979– 993 (1979).
Volknandt, W. et al. J. Biochem. 202, 855– 861 (1991).
Glaser, L., Melo, A. & Paul, R. J. Biol. Chem. 242, 1944–1954 (1967).
Neu, H.C. J. Biol. Chem. 242, 3896–3904 (1967).
Burns, D.M. & Beacham, I.R. Nucleic Acids Res. 14, 4325–4342 (1986).
Koonin, E.V. Protein Sci. 3, 356–358 (1994).
Sträter, N, Lipscomb, W.N., Klabunde, T. & Krebs, B. Angew. Chem. Int. Edn Engl. 35, 2024– 2055 (1996).
Sträter, N., Klabunde, T., Tucker, P., Witzel, H. & Krebs, B. Science 268, 1489– 1492 (1995).
Klabunde, T., Sträter, N., Fröhlich, R., Witzel, H. & Krebs, B. J. Mol. Biol. 259, 737–748 (1996).
Goldberg, J. et al. Nature 376, 745–753 (1995).
Egloff, M.-P., Cohen, P.T.W., Reinemer, P. & Barford, D. J. Mol. Biol. 254, 942–959 (1995).
Kissinger, C.R. et al. Nature 378, 641–644 (1995).
Griffith, J.P. et al. Cell 82, 507–522 (1995).
Neu, H.C. Biochemistry 7, 3766–3773 (1968).
Holm, L. & Sander, C. J. Mol. Biol. 233, 123–138 (1993).
Dvorak, H.F. & Heppel, L.A. J. Biol. Chem. 243, 2647–2653 (1968).
Klabunde, T. & Krebs, B. Struct. Bonding 89, 177–198 (1997).
Ruiz, A., Hurtado, C., Ribeiro, J.M., Sillero, A. & Sillero, M.A. G. J. Bacteriol. 171 , 6703–6709 (1989).
Otwinowski, Z. in Proceedings of the CCP4 study weekend: data collection and processing (eds Sawyer, L., Isaacs, N. & Bayley, S.) 56– 62 (SERC Daresbury Laboratory, Warrington, UK; 1993 ).
De La Fortelle, E. & Bricogne, G. Methods Enzymol. 276, 472–494 ( 1997).
CCP4. Acta Crystallogr. D 50, 760–763 (1994).
Jones, T.A., Zou, J.Y., Cowan, S. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 ( 1991).
Brünger, A.T. X–PLOR: a system for crystallography and NMR Version 3.1 (Yale University Press, New Haven, CT; 1992).
Barton, G.J. & Sternberg, M.J.E. Methods Enzymol. 183, 403–428 (1990).
Barton, G.J. & Sternberg, M.J. E. Protein Eng. 1, 89–94 (1987).
Kraulis, P.J. J. Appl. Crystallogr. 24, 946–950 (1991).
Meritt, E.A. & Murphy, M.E.P. Acta Crystallogr. D 50, 869–873 (1994).
Nicholls, A., Sharp, K.A. & Honig, B. Proteins 11, 281– 296 (1991).
Gilson, M.K., Sharp, K.A. & Honig, B.H. J. Comput. Chem. 9, 327– 335 (1987).
Esnouf, R.M. J. Mol. Graphics 15, 133–138 (1997).
Barton, G.J. Protein Eng. 6, 37–40 ( 1993).
Acknowledgements
We thank W. Saenger for generous support, G. Leonard (ESRF and EMBL, Grenoble, France) for assistance with the MAD measurements, W. Rypniewski for help with the data collection at the EMBL beamline at DESY, Hamburg, and G. Bains for support during the data collection at the ESRF and for helpful comments on the manuscript. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to N.S.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Knöfel, T., Sträter, N. X-ray structure of the Escherichia coli periplasmic 5'-nucleotidase containing a dimetal catalytic site. Nat Struct Mol Biol 6, 448–453 (1999). https://doi.org/10.1038/8253
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/8253
This article is cited by
-
Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5′-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives
Purinergic Signalling (2021)
-
Difference in NaCl tolerance of membrane-bound 5′-nucleotidases purified from deep-sea and brackish water Shewanella species
Extremophiles (2017)
-
Cellular function and molecular structure of ecto-nucleotidases
Purinergic Signalling (2012)
-
Ecto-5’-nucleotidase: Structure function relationships
Purinergic Signalling (2006)
-
Invited Lectures
Purinergic Signalling (2006)