Abstract
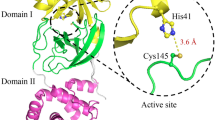
THE picornavirus family includes several pathogens such as poliovirus, rhinovirus (the major cause of the common cold), hepat-itis A virus and the foot-and-mouth disease virus. Picornaviral proteins are expressed by direct translation of the genomic RNA into a single, large polyprotein precursor1,2. Proteolysis of the viral polyprotein into the mature proteins is assured by the viral 3C enzymes, which are cysteine proteinases3–6. Here we report the X-ray crystal structure at 2.3 Å resolution of the 3C proteinase from hepatitis A virus (HAV-3C). The overall architecture of HAV-3C reveals a fold resembling that of the chymotrypsin family of serine proteinases, which is consistent with earlier predictions7,8. Cata-lytic residues include Cys 172 as nucleophile and His 44 as general base. The 3C cleavage specificity for glutamine residues is defined primarily by His 191. The overall structure suggests that an inter-molecular (trans) cleavage releases 3C and that there is an active proteinase in the polyprotein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Palmenberg, A. C. A. Rev. Microbiol. 44, 603–623 (1990).
Dougherty, W. G. & Semler, B. L. Microbiol. Rev. 57, 781–822 (1993).
Argos, P., Kamer, G., Nicklin, M. J. H. & Wimmer, E. Nucleic Acids Res. 12, 7251–7267 (1984).
Ivanoff, L. A., Towatari, T., Ray, J., Korant, B. D. & Petteway, S. R. Jr . Proc. natn. Acad. Sci. U.S.A. 83, 5392–5396 (1986).
Hämmerle, T., Hellen, C. U. T. & Wimmer, E. J. biol. Chem. 266, 5412–5416 (1991).
Kean, K. M., Teterina, N. L., Marc, D. & Girard, M. Virology 181, 609–619 (1991).
Gorbalenya, A. E., Donchenko, A. P., Blinov, V. M. & Koonin, E. V. FEBS Lett. 243, 103–114 (1989).
Bazan, J. F. & Fletterick, R. J. Proc. natn. Acad. Sci. U.S.A. 85, 7872–7876 (1988).
Chernaia, M. M., Malcolm, B. A., Allaire, M. & James, M. N. G. J. molec. Biol. 234, 890–893 (1993).
Chothia, C. & Janin, J. Biochemistry 21, 3955–3965 (1982).
Fujinaga, M., Delbaere, L. T. J., Brayer, G. D. & James, M. N. G. J. molec. Biol. 183, 479–502 (1985).
Tong, L., Wengler, G. & Rossmann, M. G. J. molec. Biol. 230, 228–247 (1993).
Fujinaga, M. et al. J. molec. Biol. 195, 397–418 (1987).
Drenth, J., Kalk, K. H. & Swen, H. M. Biochemistry 15, 3731–3738 (1976).
Kraut, J. et al. Cold Spring Harbor Symp. quant. Biol. 36, 117–123 (1971).
Jewell, D. A., Swietnicki, W., Dunn, B. M. & Malcolm, B. A. Biochemistry 31, 7862–7869 (1992).
Read, R. J. Acta. crystallogr. A42, 140–149 (1986).
Sakabe, N. Nucl. Inst. Meth. Phys. Res. A303, 448–463 (1991).
Higashi, T. J. appl. Crystallogr. 22, 9–18 (1989).
Sheldrick, G. M. Acta. crystallogr. A46, 467–473 (1990).
Otwinowski, Z. CCP4 Study Weekend 80–86 (Daresbury, UK, 1991).
Cohen, J. I., Ticehurst, J. R., Purcell, R. H., Buckler-White, A. & Baroudy, B. M. J. Virol. 61, 50–59 (1987).
Brünger, A. T. X-PLOR Version 3.1 Manual (Yale Univ. Press, New Haven and London, 1992).
Hendrickson, W. A. Meth. Enzym. 115, 252–270 (1985).
Tronrud, D. E., TenEyck, L. F. & Matthews, B. W. Acta crystallogr. A43, 489–503 (1987).
Jones, T. A. Meth. Enzym. 115, 157–171 (1985).
Morris, A. L., MacArthur, M. W., Hutchinson, E. G. & Thornton, J. M. Proteins 12, 345–364 (1992).
Lüthy, R., Bowie, J. U. & Eisenberg, D. Nature 356, 83–85 (1992).
Kraulis, P. J. J. appl. Crystallogr. 24, 946–950 (1991).
Evans, S. V. J. molec. Graphics 11, 134–138 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Allaire, M., Chernaia, M., Malcolm, B. et al. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature 369, 72–76 (1994). https://doi.org/10.1038/369072a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/369072a0
This article is cited by
-
Autoprocessing and oxyanion loop reorganization upon GC373 and nirmatrelvir binding of monomeric SARS-CoV-2 main protease catalytic domain
Communications Biology (2022)
-
The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks
Nature Microbiology (2020)
-
Conformational Flexibility of a Short Loop near the Active Site of the SARS-3CLpro is Essential to Maintain Catalytic Activity
Scientific Reports (2016)
-
Protease 3C of hepatitis A virus induces vacuolization of lysosomal/endosomal organelles and caspase-independent cell death
BMC Cell Biology (2015)
-
Three-dimensional structure of foot-and-mouth disease virus and its biological functions
Archives of Virology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.