Abstract
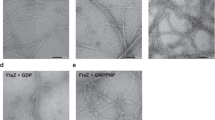
Bacterial cell division ends with septation, the constriction of the cell wall and cell membranes that leads to the formation of two daughter cells1,2. During septation, FtsZ, a protein of relative molecular mass 40,000 which is ubiquitous in eubacteria and is also found in archaea and chloroplasts3, localizes early at the division site to form a ring-shaped septum. This septum is required for the mechanochemical process of membrane constriction4. FtsZ is a GTPase5,6 with weak sequence homology to tubulins7. The nature of FtsZ polymers in vivo is unknown, but FtsZ can form tubules, sheets and minirings in vitro8,9. Here we report the crystal structure at 2.8 Å resolution of recombinant FtsZ from the hyperthermophilic methanogen Methanococcus jannaschii. FtsZ has two domains, one of which is a GTPase domain with a fold related to one found in the proteins p21ras and elongation factor EF-Tu. The carboxy-terminal domain, whose function is unknown, is a four-stranded β-sheet tilted by 90° against the β-sheet of the GTPase domain. The two domains are arranged around a central helix. GDP binding is different from that typically found in GTPases and involves four phosphate-binding loops and a sugar-binding loop in the first domain, with guanine being recognized by residues in the central connecting helix. The three-dimensional structure of FtsZ is similar to the structure of α- and β-tubulin10.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rothfield, L. I. & Justice, S. S. Bacterial cell division: the cycle of the ring. Cell 88, 581–584 (1997).
Donachie, W. D. The cell cycle of Escherichia coli. Annu. Rev. Microbiol. 47, 199–230 (1993).
Erickson, H. P. FtsZ, a tubulin homologue, in prokaryote cell division. Trends Cell Biol. 7, 362–367 (1997).
Lutkenhaus, J. FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 9, 403–409 (1993).
de Boer, P., Crossley, R. & Rothfield, L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359, 254–256 (1992).
RayChaudhuri, D. & Park, J. T. Escherichia coli cell-division gene FtsZ encodes a novel GTP-binding protein. Nature 359, 251–254 (1992).
Erickson, H. P. FtsZ, a prokaryotic homolog of tubulin? Cell 80, 367–370 (1995).
Bramhill, D. & Thompson, C. M. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl Acad. Sci. USA 91, 5813–5817 (1994).
Mukherjee, A. & Lutkenhaus, J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176, 2754–2758 (1994).
Nogales, E., Wolf, S. G. & Downing, K. H. Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 (1998).
Bult, C. J. et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 269, 496–512 (1996).
Miroux, B. & Walker, J. E. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996).
Tong, L., de Vos, A. M., Milburn, M. V. & Kim, S.-H. Crystal structures at 2.2 Å resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J. Mol. Biol. 217, 503–513 (1991).
Rossmann, M. G., Moras, D. & Olsen, K. Chemical and biological evolution of a nucleotide-binding protein. Nature 250, 194–199 (1974).
Chook, Y. M., Gray, J. V., Ke, H. & Lipscomb, W. N. The monofunctional chorismate mutase from Bacillus subtilis: structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications on the mechanism of enzymatic reaction. J. Mol. Biol. 240, 476–500 (1994).
Linse, K. & Mandelkow, E.-M. The GTP-binding peptide of β-tubulin. J. Biol. Chem. 263, 15205–15210 (1988).
Sage, C. R. et al. Site-directed mutagenesis of putative GTP-binding sites of yeast β-tubulin: evidence that α-, β- and γ-tubulins are atypical GTPases. Biochemistry 34, 16870–16875 (1995).
Bourne, H. R., Sanders, D. A. & McCormick, F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 (1991).
Mukherjee, A., Dai, K. & Lutkenhaus, J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl Acad. Sci. USA 90, 1053–1057 (1993).
de Pereda, J. M., Leynadier, D., Evangelio, J. A., Chacon, P. & Andreu, J. M. Tubulin secondary structure analysis, limited proteolysis sites, and homology to FtsZ. Biochemistry 35, 14203–14215 (1996).
Sackett, D. L. Structure and function in the tubulin dimer and the role of the acidic carboxyl terminus. Subcell. Biochem. 24, 255–302 (1995).
Leslie, A. G. W. Recent changes to the MOSFLM package for processing film and image plate data (SERC Laboratory, Daresbury, Warrington, UK, (1991)).
The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 54, 760–763 (1994).
Sheldrick, G. M. Heavy atom location using SHELXS-90.In Isomorphous Replacement and Anomalous Scattering: Proceedings of the CCP4 Study Weekend 25–26 January 1991 (eds Wolf, W., Evans, P. R. & Leslie, A. G. W.) 23–38 (SERC Daresbury Laboratory, Warrington, UK, (1991)).
Abrahams, J. P. & Leslie, A. G. W. Methods used in the structure determination of bovine mitochondrial F1-ATPase. Acta Crystallogr. D 52, 30–42 (1996).
Jones, T. A. Agraphics model building and refinement system for macromolecules. J. Appl. Cryst. 11, 268–272 (1978).
Brünger, A. X-PLOR Version 3.1, A System for X-ray Crystallography and NMR (Yale University Press, New Haven and London, (1992)).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24, 946–950 (1991).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Barton, G. J. ALSCRIPT: a tool to format multiple sequence alignments. Prot. Eng. 6, 37–40 (1993).
Acknowledgements
We thank H. Huber (University of Regensburg) for M. jannaschii cells, I. Fearnley for mass spectrometry, B. Miroux for C41 cells and for discussion, and A. Murzin for pointing out the similarity to chorismate mutase. J.L. is supported by an EMBO long-term fellowship.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Löwe, J., Amos, L. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 (1998). https://doi.org/10.1038/34472
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/34472
This article is cited by
-
Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody
Nature Communications (2023)
-
Mycobacterial FtsZ and inhibitors: a promising target for the anti-tubercular drug development
Molecular Diversity (2023)
-
The mycotoxin viriditoxin induces leukemia- and lymphoma-specific apoptosis by targeting mitochondrial metabolism
Cell Death & Disease (2022)
-
Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism
Nature Communications (2021)
-
FtsZ: The Force Awakens
Journal of the Indian Institute of Science (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.