Abstract
Notch proteins are ligand-activated transmembrane receptors involved in cell-fate selection throughout development1,2,3. No known enzymatic activity is contained within Notch and the molecular mechanism by which it transduces signals across the cell membrane is poorly understood. In many instances, Notch activation results in transcriptional changes in the nucleus through an association with members of the CSL family of DNA-binding proteins (where CSL stands for CBF1, Su(H), Lag-1)1,2,3,4. As Notch is located in the plasma membrane and CSL is a nuclear protein, two models have been proposed to explain how they interact (Fig. 1) . The first suggests that the two interact transiently at the membrane1,5,6,7. The second postulates that Notch is cleaved by a protease, enabling the cleaved fragment to enter the nucleus6,8,9,10,11,12,13,14. Here we show that signalling by a constitutively active membrane-bound Notch-1 protein requires the proteolytic release of the Notch intracellular domain (NICD), which interacts preferentially with CSL. Very small amounts of NICD are active, explaining why it is hard to detect in the nucleus in vivo. We also show that it is ligand binding that induces release of NICD.

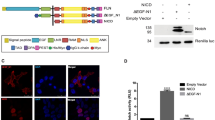
a, Models not invoking processing propose that interactions between Notch and CSL at the membrane may be sufficient to transduce a signal. Thus, ligand-regulated NICD release is not expected to be necessary for signalling. b, The processing model suggests that Notch signalling requires the release of NICD, which is capable of direct interaction with CSL in the nucleus, to turn on transcription of target genes. This model predicts that NICD release is regulated by ligand binding and that blocking proteolysis interferes with signalling. Notch-1 domain symbols as in Fig. 2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Artavanis-Tsakonas, S., Matsuno, K. & Fortini, M. E. Notch signaling. Science 268, 225–232 (1995).
Kopan, R. & Turner, D. The Notch pathway: democracy and aristrocracy during the selection of cell fate. Curr. Opin. Neurobiol. 6, 594–601 (1996).
Weinmaster, G. The ins and outs of Notch signaling. Mol. Cell. Neurosci. 9, 91–102 (1997).
Artavanis-Tsakonas, S. Alagille syndrome—a notch up for the Notch receptor. Nature Genet. 16, 212–213 (1997).
Fortini, M. E. & Artavanis-Tsakonas, S. The suppressor of hairless protein participates in Notch receptor signaling. Cell 79, 273–282 (1994).
Roehl, H., Bosenberg, M., Blelloch, R. & Kimble, J. Roles of the Ram and Ank domains in signaling by the C-elegans Glp-1 receptor. EMBO J. 15, 7002–7012 (1996).
Aster, J. C. et al. Oncogenic forms of Notch1 lacking either the primary binding site for Rbp-J-kappa or nuclear localization sequences retain the ability to associate with Rbp-J-kappa and activate transcription. J. Biol. Chem. 272, 11336–11343 (1997).
Lieber, T., Kidd, S., Alcamo, E., Corbin, V. & Young, M. W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 7, 1949–1965 (1993).
Kopan, R., Nye, J. S. & Weintraub, H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development 120, 2385–2396 (1994).
Hsieh, J. J.-D. & Hayward, S. D. Masking of the CBF1/RBPjκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science 268, 560–563 (1995).
Jarriault, S. et al. Signalling downstream of activated mammalian Notch. Nature 377, 355–358 (1995).
Tamura, K. et al. Physical interaction between a novel domain of the receptor Notch and the transcription factor Rbp-J-kappa/Su(H). Curr. Biol. 5, 1416–1423 (1995).
Kopan, R., Schroeter, E. H., Weintraub, H. & Nye, J. S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl Acad. Sci. USA 93, 1683–1687 (1996).
Wettstein, D. A., Turner, D. L. & Kintner, C. The Xenopus homolog of Drosophila Suppressor Of Hairless mediates Notch signaling during primary neurogenesis. Development 124, 693–702 (1997).
Bredenbeek, P. J., Frolov, I., Rice, C. M. & Schlesinger, S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67, 6439–6446 (1993).
Fujiwara, T., Oda, K., Yokota, S., Takatsuki, A. & Ikehara, Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 263, 18545–18552 (1988).
Doms, R. W., Russ, G. & Yewdell, J. W. Brefeldin A redistributes residence and itinerant Golgi proteins to the endoplasmic reticulum. J. Cell Biol. 109, 61–72 (1989).
Griffiths, G., Quinn, P. & Warren, G. Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus. J. Cell Biol. 96, 835–850 (1983).
Quinn, P., Griffiths, G. & Warren, G. Dissection of the Golgi complex. II. Density separation of specific Golgi functions in virally infected cells treated with monensin. J. Cell Biol. 96, 851–856 (1983).
Pan, D. J. & Rubin, G. M. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90, 271–280 (1997).
1. Sotillos, S., Roch, F. & Campuzano, S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124, 4769–4779 (1997).
Wen, C., Metzstein, M. M. & Greenwald, I. Sup-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in the Lin-12/Notch signaling. Development 124, 4759–4767 (1997).
Blaumueller, C. M., Qi, H. L., Zagouras, P. & Artavanis-Tsakonas, S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90, 281–291 (1997).
Gho, M., Lecourtois, M., Geraud, G., Posakony, J. W. & Schweisguth, F. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 122, 1673–1682 (1996).
Honjo, T. The shortest path from the surface to the nucleus: RBPjk/Su(H) transcription factor. Genes Cells 1, 1–9 (1996).
Weintraub, H. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc. Natl Acad. Sci. USA 85, 5819–5823 (1988).
Lindsell, C. E., Shawber, C. J., Boulter, J. & Weinmaster, G. Jagged: a mammalian ligand that activates Notch1. Cell 80, 909–917 (1995).
Capobianco, A. J., Zagouras, P., Blaumueller, C. M., Artavanis-Tsakonas, S. & Bishop, J. M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17, 6265–6273 (1997).
Brown, M. S. & Goldstein, J. L. The SREBP pathway—regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997).
Turner, D. L. & Weintraub, H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434–1447 (1994).
Acknowledgements
We thank A. Israel for providing us with the FLAG–CSLRBP3 expression vector; I.Frolov for the Sindbis virus vectors pSINrep5 and DHBB; G. Rubin and D. J. Pan for the Kuzbanian cDNAs; D. Syder and M. Crankshaw for technical help; R. Cagan, I. Boime, J. Gordon, D. Ornitz and members of the Kopan lab for reading and commenting on the manuscript; and G. Goldberg, G. Grant and B. Marmer for discussions and technical advice. R. K. Thanks H. Weintraub for encouragement and support. This research was supported by the NIH.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schroeter, E., Kisslinger, J. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998). https://doi.org/10.1038/30756
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/30756
This article is cited by
-
NOTCH localizes to mitochondria through the TBC1D15-FIS1 interaction and is stabilized via blockade of E3 ligase and CDK8 recruitment to reprogram tumor-initiating cells
Experimental & Molecular Medicine (2024)
-
Soluble and multivalent Jag1 DNA origami nanopatterns activate Notch without pulling force
Nature Communications (2024)
-
Notch intracellular domains form transcriptionally active heterodimeric complexes on sequence-paired sites
Scientific Reports (2024)
-
Ubiquitin-specific protease 28: the decipherment of its dual roles in cancer development
Experimental Hematology & Oncology (2023)
-
p63: a crucial player in epithelial stemness regulation
Oncogene (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.