Abstract
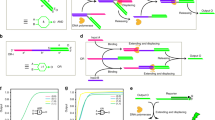
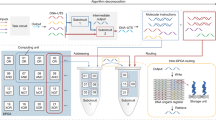
DNA is a reliable biomolecule with which to build molecular computation systems. In particular, DNA logic circuits (diffusion-based) have shown good performance regarding scalability and correctness of computation. However, previous architectures of DNA logic circuits have two limitations. First, the speed of computation is slow, often requiring hours to compute a simple function. Second, the circuits are of high complexity regarding the number of DNA strands. Here, we introduce an architecture of DNA logic circuits based on single-stranded logic gates using strand-displacing DNA polymerase. The logic gates consist of only single DNA strands, which largely reduces leakage reactions and signal restoration steps such that the circuits are improved in regard to both speed of computation and the number of DNA strands needed. Large-scale logic circuits can be constructed from the gates by simple cascading strategies. In particular, we have demonstrated a fast and compact logic circuit that computes the square-root function of four-bit input numbers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data used in this paper are available from the corresponding author upon proper request.
References
Watson, J. D. & Crick, F. H. Molecular structure of nucleic acids. Nature 171, 737–738 (1953).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Qian, L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011).
Chatterjee, G., Dalchau, N., Muscat, R. A., Phillips, A. & Seelig, G. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechnol. 12, 920 (2017).
Srinivas, N., Parkin, J., Seelig, G., Winfree, E. & Soloveichik, D. Enzyme-free nucleic acid dynamical systems. Science 358, eaal2052 (2017).
Cherry, K. M. & Qian, L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 559, 370 (2018).
Seelig, G., Soloveichik, D., Zhang, D. Y. & Winfree, E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Thubagere, A. J. et al. Compiler-aided systematic construction of large-scale DNA strand displacement circuits using unpurified components. Nat. Commun. 8, 14373 (2017).
Bui, H. et al. Localized DNA hybridization chain reactions on DNA origami. ACS Nano 12, 1146–1155 (2018).
Wang, B., Thachuk, C., Ellington, A. D., Winfree, E. & Soloveichik, D. Effective design principles for leakless strand displacement systems. Proc. Natl Acad. Sci. USA 115, E12182–E12191 (2018).
Thachuk, C., Winfree, E. & Soloveichik, D. in International Workshop on DNA-Based Computers Vol. 9211 (eds Phillips, A. & Yin, P.) 133–153 (Springer, 2015).
Boneh, D., Dunworth, C., Lipton, R. J. & Sgall, J. On the computational power of DNA. Discrete Appl. Math. 71, 79–94 (1996).
Ogihara, M. & Ray, A. Simulating boolean circuits on a DNA computer. Algorithmica 25, 239–250 (1999).
Winfree, E. Whiplash PCR for O(1) Computing (California Institute of Technology, 1998).
Baccouche, A., Montagne, K., Padirac, A., Fujii, T. & Rondelez, Y. Dynamic DNA-toolbox reaction circuits: a walkthrough. Methods 67, 234–249 (2014).
Montagne, K., Plasson, R., Sakai, Y., Fujii, T. & Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011).
Padirac, A., Fujii, T. & Rondelez, Y. Bottom-up construction of in vitro switchable memories. Proc. Natl Acad. Sci. USA 109, E3212–E3220 (2012).
Aubert, N., Mosca, C., Fujii, T., Hagiya, M. & Rondelez, Y. Computer-assisted design for scaling up systems based on DNA reaction networks. J. R. Soc. Interface 11, 20131167 (2014).
van Roekel, H. W. et al. Programmable chemical reaction networks: emulating regulatory functions in living cells using a bottom-up approach. Chem. Soc. Rev. 44, 7465–7483 (2015).
Yordanov, B. et al. Computational design of nucleic acid feedback control circuits. ACS Synth. Biol. 3, 600–616 (2014).
Kishi, J. Y., Schaus, T. E., Gopalkrishnan, N., Xuan, F. & Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 10, 155 (2018).
Acknowledgements
This work is supported by NSF Grants nos. CCF-1320360, CCF-1217457, CCF-1617791 and CCF-1813805.
Author information
Authors and Affiliations
Contributions
T.S. and J.R. designed the logic gates and circuits. T.S. designed and performed the experiments. T.S. analysed the data. T.S. and J.R. wrote the paper. A.E., S.S., H.B., D.F., M.Y. and R.M. contributed to creating the logic gates and circuits. All authors discussed the design and results of the experiments. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary Sections 1–5, Figs. 1–7, Tables 1–13 and Ref. 1.
Rights and permissions
About this article
Cite this article
Song, T., Eshra, A., Shah, S. et al. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat. Nanotechnol. 14, 1075–1081 (2019). https://doi.org/10.1038/s41565-019-0544-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-019-0544-5
This article is cited by
-
Controllable and reusable seesaw circuit based on nicking endonucleases
Journal of Nanobiotechnology (2024)
-
Heteromultivalency enables enhanced detection of nucleic acid mutations
Nature Chemistry (2024)
-
DNA as a universal chemical substrate for computing and data storage
Nature Reviews Chemistry (2024)
-
Characterization of Cascaded DNA Generation Reaction for Amplifying DNA Signal
New Generation Computing (2024)
-
The structural power of reconfigurable circuits in the amoebot model
Natural Computing (2024)