Abstract
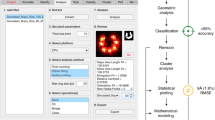
This protocol describes the reconstruction of biological molecules from the electron micrographs of single particles. Computation here is performed using the image-processing software SPIDER and can be managed using a graphical user interface, termed the SPIDER Reconstruction Engine. Two approaches are described to obtain an initial reconstruction: random-conical tilt and common lines. Once an existing model is available, reference-based alignment can be used, a procedure that can be iterated. Also described is supervised classification, a method to look for homogeneous subsets when multiple known conformations of the molecule may coexist.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
































Similar content being viewed by others
References
Frank, J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies (Oxford University Press, New York, 2006).
Glaeser, R.M., Downing, K., DeRosier, D., Chiu, W. & Frank, J. Electron Crystallography of Biological Macromolecules (Oxford University Press, New York, 2007).
Frank, J., Shimkin, B. & Dowse, H. SPIDER-a modular software system for electron image processing. Ultramicroscopy 6, 343–358 (1981).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
van Heel, M. & Keegstra, W. Imagic: a fast, flexible, and friendly image processing software system. Ultramicroscopy 7, 113–129 (1981).
van Heel, M., Harauz, G. & Orlova, E.V. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Marabini, R. et al. Xmipp: and image processing package for electron microscopy. J. Struct. Biol. 116, 237–240 (1996).
Aebi, U., Carragher, B. & Smith, P.R. Editorial. J. Struct. Biol. 116, 1 (1996).
Schoehn, G. et al. An archaeal peptidase assembles into two different quaternary structures: A tetrahedron and a giant octahedron. J. Biol. Chem. 281, 36327–36337 (2006).
Halic, M. et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444, 507–511 (2006).
Taylor, D.J. et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 26, 2421–2431 (2007).
Frank, J., Goldfarb, W., Eisenberg, D. & Baker, T.S. Reconstruction of glutamine synthetase using computer averaging. Ultramicroscopy 3, 283–290 (1978).
Radermacher, M., Wagenknecht, T., Verschoor, A. & Frank, J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli . J. Microsc. 146, 113–136 (1987).
Radermacher, M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J. Electron Microsc. Tech. 9, 359–394 (1988).
Qazi, U., Gettins, P.G.W. & Stoops, J.K. On the structural changes of native human α2-macroglobulin upon proteinase entrapment. Three-dimensional structure of the half-transformed molecule. J. Biol. Chem. 273, 8987–8993 (1998).
Radermacher, M. et al. The three-dimensional structure of complex I from Yarrowia lipolytica: a highly dynamic enzyme. J. Struct. Biol. 154, 269–279 (2006).
Ohi, M.D., Ren, L., Wall, J.S., Gould, K.L. & Walz, T. Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc. Natl. Acad. Sci. USA 104, 3195–3200 (2007).
Andel, F., Ladurner, A.G., Inouye, C., Tjian, R. & Nogales, E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286, 2153–2156 (1999).
Craighead, J.L., Chang, W.H. & Asturias, F.J. Structure of yeast RNA polymerase II in solution: implications for enzyme regulation and interaction with promoter DNA. Structure 10, 1117–1125 (2002).
van Heel, M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21, 111–124 (1987).
Penczek, P.A., Zhu, J. & Frank, J. A common-lines based method for determining orientations for N>3 particle projections simultaneously. Ultramicroscopy 63, 205–218 (1996).
Crowther, R.A., DeRosier, D.J. & Klug, A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc. Roy. Soc. Lond. A. 317, 319–340 (1970).
Gabashvili, I.S. et al. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100, 537–549 (2000).
Valle, M. et al. Cryo-EM reveals an active role for the aminoacyl-tRNA in the accommodation process. EMBO J. 21, 3557–3567 (2002).
Gao, H., Valle, M., Ehrenberg, M. & Frank, J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J. Struct. Biol. 147, 283–290 (2004).
Frigo, M. & Johnson, S.G. FFTW: an adaptive software architecture for the FFT. Vol. 3, 1381–1384 (23rd International Conference on Acoustics, Speech, and Signal Processing; Proc. ICASSP, Seattle, 1998).
Baxter, W.T., Leith, A. & Frank, J. SPIRE: the SPIDER reconstruction engine. J. Struct. Biol. 157, 56–63 (2007).
Mouche, F., Boisset, N. & Penczek, P.A. Lumbricus terrestris hemoglobin—the architecture of linker chains and structural variation of the central toroid. J. Struct. Biol. 133, 176–192 (2001).
Scheres, S.H. et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods 4, 27–29 (2007).
Penczek, P., Radermacher, M. & Frank, J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy 40, 33–53 (1992).
van Heel, M. & Frank, J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6, 187–194 (1981).
Boisset, N., Penczek, P., Pochon, F., Frank, J. & Lamy, J. Three-dimensional architecture of human alpha 2-macroglobulin transformed with methylamine. J. Mol. Biol. 232, 522–529 (1993).
Roseman, A.M. Particle finding in electron micrographs using a fast local correlation algorithm. Ultramicroscopy 94, 225–236 (2003).
Rath, B.K. & Frank, J. Fast automatic particle picking from cryo-electron micrographs using a locally normalized cross-correlation function: a case study. J. Struct. Biol. 145, 84–90 (2004).
Zhu, J., Penczek, P.A., Schröder, R. & Frank, J. Three-dimensional reconstruction with contrast transfer function correction from energy-filtered cryoelectron micrographs: procedure and application to the 70S Escherichia coli ribosome. J. Struct. Biol. 118, 197–219 (1997).
Acknowledgements
This article is dedicated to the memory of our good friend and colleague Nicolas Boisset, who passed away on January 4, 2008. The authors would like to thank Jesse Brown for batch files on the common-lines approach and helpful discussions. We also thank Michael Watters for assistance with the preparation of the illustrations. Supported by HHMI and NIH grants P41 RR01219 and R37 GM29169 (to J.F.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
41596_2008_BFnprot2008156_MOESM350_ESM.pdf
Supplementary Fig. 3. JWEB window after clicking Fit Angles. The tilt angle theta, and tilt-axis directions phi and gamma have been calculated. (PDF 217 kb)
Rights and permissions
About this article
Cite this article
Shaikh, T., Gao, H., Baxter, W. et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3, 1941–1974 (2008). https://doi.org/10.1038/nprot.2008.156
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.156
This article is cited by
-
Structural insights into the binding mechanism of Clr4 methyltransferase to H3K9 methylated nucleosome
Scientific Reports (2024)
-
Structural mechanism of mitochondrial membrane remodelling by human OPA1
Nature (2023)
-
Disome-seq reveals widespread ribosome collisions that promote cotranslational protein folding
Genome Biology (2021)
-
Scanning probe microscopy
Nature Reviews Methods Primers (2021)
-
In situ structure and organization of the influenza C virus surface glycoprotein
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.