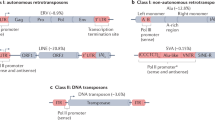
Abstract
Transposable elements are DNA segments with the unique ability to move about in the genome. This inherent feature can be exploited to harness these elements as gene vectors for genome manipulation. Transposon-based genetic strategies have been established in vertebrate species over the last decade, and current progress in this field suggests that transposable elements will serve as indispensable tools. In particular, transposons can be applied as vectors for somatic and germline transgenesis, and as insertional mutagens in both loss-of-function and gain-of-function forward mutagenesis screens. In addition, transposons will gain importance in future cell-based clinical applications, including nonviral gene transfer into stem cells and the rapidly developing field of induced pluripotent stem cells. Here we provide an overview of transposon-based methods used in vertebrate model organisms with an emphasis on the mouse system and highlight the most important considerations concerning genetic applications of the transposon systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
11 June 2009
NOTE: In the version of this article initially published, a part of Figure 1b was incorrectly labeled. The error has been corrected in the HTML and PDF versions of the article.
References
Feng, Q., Moran, J.V., Kazazian, H.H., Jr. & Boeke, J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 (1996).
Mathias, S.L., Scott, A.F., Kazazian, H.H. Jr., Boeke, J.D. & Gabriel, A. Reverse transcriptase encoded by a human transposable element. Science 254, 1808–1810 (1991).
Luan, D.D., Korman, M.H., Jakubczak, J.L. & Eickbush, T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605 (1993).
Cost, G.J., Feng, Q., Jacquier, A. & Boeke, J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910 (2002).
Zwaal, R.R., Broeks, A., van Meurs, J., Groenen, J.T. & Plasterk, R.H. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl. Acad. Sci. USA 90, 7431–7435 (1993).
Thibault, S.T. et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283–287 (2004). Demonstration that full genome coverage in Drosophila cannot be done by using a single transposon owing to distinct preferences for genomic insertion sites.
Ivics, Z., Hackett, P.B., Plasterk, R.H. & Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 (1997). First report on the resurrection of an extinct transposon and the first class II transposon ever reported to jump in a vertebrate cell.
Mates, L., Izsvak, Z. & Ivics, Z. Technology transfer from worms and flies to vertebrates: transposition-based genome manipulations and their future perspectives. Genome Biol. 8 (Suppl. 1), S1 (2007).
Izsvak, Z., Ivics, Z. & Plasterk, R.H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 302, 93–102 (2000).
Zayed, H., Izsvak, Z., Walisko, O. & Ivics, Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol. Ther. 9, 292–304 (2004).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).Report describes efficient transposition of the insect piggyBac transposon in mice.
Koga, A. et al. The Tol1 element of medaka fish is transposed with only terminal regions and can deliver large DNA fragments into the chromosomes. J. Hum. Genet. 52, 1026–1030 (2007).
Balciunas, D. et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169 (2006).
Urasaki, A., Morvan, G. & Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 (2006).
Kaufman, C.D., Izsvak, Z., Katzer, A. & Ivics, Z. Frog Prince transposon-based RNAi vectors mediate efficient gene knockdown in human cells. J. RNAi Gene Silencing 1, 97–104 (2005).
Ivics, Z. & Izsvak, Z. Transposons for gene therapy! Curr. Gene Ther. 6, 593–607 (2006).
Mátés, L. et al. Molecular evolution of a hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. advance online publication, doi: 10.1038/ng.343 (3 May 2009). Report of a hyperactive Sleeping Beauty transposase that mediates stable gene transfer at an efficiency similar to that of viral vectors in primary human stem cells and in mice.
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Cadinanos, J. & Bradley, A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 35, e87 (2007).
Woltjen, K. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 (2009). This and the next paper show how to use the piggyBac transposon as a vector for iPSC reprogramming.
Yusa, K., Rad, R., Takeda, J. & Bradley, A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods 6, 363–369 (2009).
Henikoff, S. Conspiracy of silence among repeated transgenes. Bioessays 20, 532–535 (1998).
Sasakura, Y., Awazu, S., Chiba, S. & Satoh, N. Germ-line transgenesis of the Tc1/mariner superfamily transposon Minos in Ciona intestinalis. Proc. Natl. Acad. Sci. USA 100, 7726–7730 (2003).
Koga, A., Cheah, F.S., Hamaguchi, S., Yeo, G.H. & Chong, S.S. Germline transgenesis of zebrafish using the medaka Tol1 transposon system. Dev. Dyn. 237, 2466–2474 (2008).
Kawakami, K., Shima, A. & Kawakami, N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97, 11403–11408 (2000).
Hamlet, M.R. et al. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis 44, 438–445 (2006).
Davidson, A.E. et al. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev. Biol. 263, 191–202 (2003).
Hermanson, S., Davidson, A.E., Sivasubbu, S., Balciunas, D. & Ekker, S.C. Sleeping Beauty transposon for efficient gene delivery. Methods Cell Biol. 77, 349–362 (2004).
Sinzelle, L. et al. Generation of trangenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 15, 751–760 (2006).
Grabher, C. et al. Transposon-mediated enhancer trapping in medaka. Gene 322, 57–66 (2003).
Sato, Y. et al. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev. Biol. 305, 616–624 (2007).
Dupuy, A.J. et al. Mammalian germ-line transgenesis by transposition. Proc. Natl. Acad. Sci. USA 99, 4495–4499 (2002).
Wilber, A. et al. RNA as a source of transposase for sleeping beauty-mediated gene insertion and expression in somatic cells and tissues. Mol. Ther. 13, 625–630 (2006).
Carlson, C.M., Frandsen, J.L., Kirchhof, N., McIvor, R.S. & Largaespada, D.A. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl. Acad. Sci. USA 102, 17059–17064 (2005).
Luo, G., Ivics, Z., Izsvak, Z. & Bradley, A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 95, 10769–10773 (1998).
Luo, G. et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26, 424–429 (2000).
Yusa, K. et al. Genome-wide phenotype analysis in ES cells by regulated disruption of Bloom's syndrome gene. Nature 429, 896–899 (2004).
Wang, W. et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 105, 9290–9295 (2008).
Wang, W., Bradley, A. & Huang, Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res (2009).
Hansen, G.M. et al. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 18, 1670–1679 (2008).
Takeda, J., Izsvak, Z. & Ivics, Z. Insertional mutagenesis of the mouse germline with Sleeping Beauty transposition. Methods Mol. Biol. 435, 109–125 (2008).
Dupuy, A.J., Fritz, S. & Largaespada, D.A. Transposition and gene disruption in the male germline of the mouse. Genesis 30, 82–88 (2001).
Horie, K. et al. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98, 9191–9196 (2001).
Horie, K. et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23, 9189–9207 (2003).
Carlson, C.M. et al. Transposon mutagenesis of the mouse germline. Genetics 165, 243–256 (2003).
Fischer, S.E., Wienholds, E. & Plasterk, R.H. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98, 6759–6764 (2001).
Kitada, K. et al. Transposon-tagged mutagenesis in the rat. Nat. Methods 4, 131–133 (2007).
Lu, B. et al. Generation of rat mutants using a coat color-tagged Sleeping Beauty transposon system. Mamm. Genome 18, 338–346 (2007).
Yae, K. et al. Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol. Cell. Biol. 26, 6185–6196 (2006).
Geurts, A.M. et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2, e156 (2006).
Keng, V.W. et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat. Methods 2, 763–769 (2005). This paper shows that local hopping of the Sleeping Beauty transposon can be applied for chromosomal region–specific saturation mutagenesis in mice.
Drabek, D. et al. Transposition of the Drosophila hydei Minos transposon in the mouse germ line. Genomics 81, 108–111 (2003).
Zagoraiou, L. et al. In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues. Proc. Natl. Acad. Sci. USA 98, 11474–11478 (2001).
Wu, S., Ying, G., Wu, Q. & Capecchi, M.R. Toward simpler and faster genome-wide mutagenesis in mice. Nat. Genet. 39, 922–930 (2007).
Kawakami, K. & Noda, T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics 166, 895–899 (2004).
Kawakami, K. et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144 (2004).
Parinov, S., Kondrichin, I., Korzh, V. & Emelyanov, A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231, 449–459 (2004).
Sivasubbu, S. et al. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech. Dev. 123, 513–529 (2006).
Balciunas, D. et al. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 5, 62 (2004).
An, W. et al. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl. Acad. Sci. USA 103, 18662–18667 (2006).
An, W. et al. Conditional activation of a single-copy L1 transgene in mice by Cre. Genesis 46, 373–383 (2008).
Collier, L.S., Carlson, C.M., Ravimohan, S., Dupuy, A.J. & Largaespada, D.A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276 (2005). This and the following paper demonstrated that the Sleeping Beauty transposon can be used as an efficient somatic mutagen to drive tumorigenesis in mice.
Dupuy, A.J., Akagi, K., Largaespada, D.A., Copeland, N.G. & Jenkins, N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005).
Largaespada, D.A. & Collier, L.S. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods Mol. Biol. 435, 95–108 (2008).
Su, Q. et al. A DNA transposon-based approach to validate oncogenic mutations in the mouse. Proc. Natl. Acad. Sci. USA 105, 19904–19909 (2008).
Keng, V.W. et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 27, 264–274 (2009).
Starr, T.K. et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323, 1747–1750 (2009).
Aitman, T.J. et al. Progress and prospects in rat genetics: a community view. Nat. Genet. 40, 516–522 (2008).
Park, I.H. et al. Disease-specific induced pluripotent stem cells. Cell 134, 877–886 (2008).
Yant, S.R. et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 25, 2085–2094 (2005).
Miskey, C., Izsvak, Z., Plasterk, R.H. & Ivics, Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31, 6873–6881 (2003).
Miskey, C. et al. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell Biol. 27, 4589–4600 (2007).
Clark, K.J., Carlson, D.F., Leaver, M.J., Foster, L.K. & Fahrenkrug, S.C. Passport, a native Tc1 transposon from flatfish, is functionally active in vertebrate cells. Nucleic Acids Res. 37, 1239–1247 (2009).
Wilson, M.H., Coates, C.J. & George, A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 15, 139–145 (2007).
Emelyanov, A., Gao, Y., Naqvi, N.I. & Parinov, S. Trans-kingdom transposition of the maize dissociation element. Genetics 174, 1095–1104 (2006).
Sinzelle, L. et al. Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc. Natl. Acad. Sci. USA 105, 4715–4720 (2008).
Han, J.S. & Boeke, J.D. A highly active synthetic mammalian retrotransposon. Nature 429, 314–318 (2004). The base composition of an L1 element was altered leading to highly efficient retrotransposition in mammalian cells.
Cost, G.J. & Boeke, J.D. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37, 18081–18093 (1998).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 4824 kb)
Rights and permissions
About this article
Cite this article
Ivics, Z., Li, M., Mátés, L. et al. Transposon-mediated genome manipulation in vertebrates. Nat Methods 6, 415–422 (2009). https://doi.org/10.1038/nmeth.1332
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1332
This article is cited by
-
First full views of a CRISPR-guided system for gene insertion
Nature (2023)
-
Gene expression changes during the evolution of the tetrapod limb
Biologia Futura (2022)
-
Exploring liver cancer biology through functional genetic screens
Nature Reviews Gastroenterology & Hepatology (2021)
-
CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma
Gene Therapy (2021)
-
A selectable, plasmid-based system to generate CRISPR/Cas9 gene edited and knock-in mosquito cell lines
Scientific Reports (2021)