Abstract
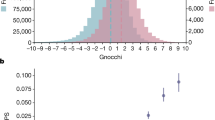
Noncoding genetic variants are likely to influence human biology and disease, but recognizing functional noncoding variants is difficult. Approximately 3% of noncoding sequence is conserved among distantly related mammals1,2,3,4, suggesting that these evolutionarily conserved noncoding regions (CNCs) are selectively constrained and contain functional variation. However, CNCs could also merely represent regions with lower local mutation rates. Here we address this issue and show that CNCs are selectively constrained in humans by analyzing HapMap genotype data. Specifically, new (derived) alleles of SNPs within CNCs are rarer than new alleles in nonconserved regions (P = 3 × 10−18), indicating that evolutionary pressure has suppressed CNC-derived allele frequencies. Intronic CNCs and CNCs near genes show greater allele frequency shifts, with magnitudes comparable to those for missense variants. Thus, conserved noncoding variants are more likely to be functional. Allele frequency distributions highlight selectively constrained genomic regions that should be intensively surveyed for functionally important variation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Thomas, J.W. et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424, 788–793 (2003).
Dermitzakis, E.T. et al. Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs). Science 302, 1033–1035 (2003).
Dermitzakis, E.T., Reymond, A. & Antonarakis, S.E. Conserved non-genic sequences – an unexpected feature of mammalian genomes. Nat. Rev. Genet. 6, 151–157 (2005).
Loots, G.G. et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288, 136–140 (2000).
Woolfe, A. et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7 (2005).
Frazer, K.A. et al. Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res. 14, 367–372 (2004).
Nobrega, M.A., Zhu, Y., Plajzer-Frick, I., Afzal, V. & Rubin, E.M. Megabase deletions of gene deserts result in viable mice. Nature 431, 988–993 (2004).
The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933 (2001).
Kryukov, G.V., Schmidt, S. & Sunyaev, S. Small fitness effect of mutations in highly conserved non-coding regions. Hum. Mol. Genet. 14, 2221–2229 (2005).
Fay, J.C., Wyckoff, G.J. & Wu, C.I. Positive and negative selection on the human genome. Genetics 158, 1227–1234 (2001).
International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Maruyama, T. & Fuerst, P.A. Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111, 675–689 (1985).
Marth, G.T., Czabarka, E., Murvai, J. & Sherry, S.T. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics 166, 351–372 (2004).
Keightley, P.D., Kryukov, G.V., Sunyaev, S., Halligan, D.L. & Gaffney, D.J. Evolutionary constraints in conserved nongenic sequences of mammals. Genome Res. 15, 1373–1378 (2005).
Cargill, M. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22, 231–238 (1999).
Charlesworth, B., Morgan, M.T. & Charlesworth, D. The effect of deleterious mutations on neutral molecular variation. Genetics 134, 1289–1303 (1993).
Dermitzakis, E.T. et al. Comparison of human chromosome 21 conserved nongenic sequences (CNGs) with the mouse and dog genomes shows that their selective constraint is independent of their genic environment. Genome Res. 14, 852–859 (2004).
Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005).
Bejerano, G. et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004).
Margulies, E.H., Blanchette, M., Haussler, D. & Green, E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 13, 2507–2518 (2003).
Keightley, P.D., Lercher, M.J. & Eyre-Walker, A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 3, e42 (2005).
Hirakawa, M. et al. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 30, 158–162 (2002).
Botstein, D. & Risch, N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 33 (Suppl.), 228–237 (2003).
Beysen, D. et al. Deletions involving long-range conserved nongenic sequences upstream and downstream of FOXL2 as a novel disease-causing mechanism in blepharophimosis syndrome. Am. J. Hum. Genet. 77, 205–218 (2005).
Emison, E.S. et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434, 857–863 (2005).
Xie, X. et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434, 338–345 (2005).
King, D.C. et al. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 15, 1051–1060 (2005).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Zar, J.H. Biostatistical Analysis 4th edn. (Prentice Hall, Upper Saddle River, New Jersey, 1999).
Acknowledgements
The authors wish to thank the International HapMap investigators, T.S. Mikkelsen for assistance determining derived alleles from chimp sequence comparisons, T. Bersaglieri for assistance genotyping SNPs, G. Rockwell for assistance implementing several statistical methods and analysis algorithms, A. Langane for DNA samples and M. Gagnebin and C. Rossier for sequencing. J.N.H. is a recipient of a Burroughs Wellcome Career Award in Biomedical Sciences, which supported this work. E.T.D. is supported by the Wellcome Trust and NIH. D.J.T. was supported by grants from the National Human Genome Research Institute. S.E.A. is supported by the Swiss National Science Foundation, NIH and EU. C.N.C. is supported by the GlaxoSmithKline Competitive Grants Award Program for Young Investigators and by a National Heart, Lung, and Blood Institute Mentored Patient Oriented Research Career Development Award.
AUTHORS' CONTRIBUTIONS
J.A.D., C.B., J.N. and D.J.T. contributed equally to this manuscript, and E.T.D. and J.N.H. co-directed this project.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Comparisons of DAF distributions, with significance levels and percentages of SNPs in DAF frequency bins. (PDF 112 kb)
Supplementary Table 2
Positions and derived allele frequencies of SNPs from dbSNP located in ultraconserved elements. (PDF 45 kb)
Rights and permissions
About this article
Cite this article
Drake, J., Bird, C., Nemesh, J. et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet 38, 223–227 (2006). https://doi.org/10.1038/ng1710
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1710
This article is cited by
-
Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci
Nature Communications (2024)
-
Genomic distribution and polymorphism of G-quadruplex motifs occupying ovine promoters and enhancers
Mammalian Genome (2023)
-
Perfect and imperfect views of ultraconserved sequences
Nature Reviews Genetics (2022)
-
MCL1 alternative polyadenylation is essential for cell survival and mitochondria morphology
Cellular and Molecular Life Sciences (2022)
-
Ultraconserved enhancer function does not require perfect sequence conservation
Nature Genetics (2021)