Abstract
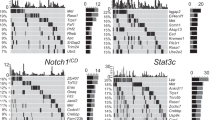
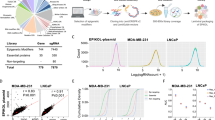
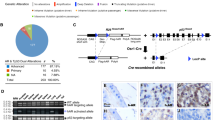
The overwhelming number of genetic alterations identified through cancer genome sequencing requires complementary approaches to interpret their significance and interactions. Here we developed a novel whole-body insertional mutagenesis screen in mice, which was designed for the discovery of Pten-cooperating tumor suppressors. Toward this aim, we coupled mobilization of a single-copy inactivating Sleeping Beauty transposon to Pten disruption within the same genome. The analysis of 278 transposition-induced prostate, breast and skin tumors detected tissue-specific and shared data sets of known and candidate genes involved in cancer. We validated ZBTB20, CELF2, PARD3, AKAP13 and WAC, which were identified by our screens in multiple cancer types, as new tumor suppressor genes in prostate cancer. We demonstrated their synergy with PTEN in preventing invasion in vitro and confirmed their clinical relevance. Further characterization of Wac in vivo showed obligate haploinsufficiency for this gene (which encodes an autophagy-regulating factor) in a Pten-deficient context. Our study identified complex PTEN-cooperating tumor suppressor networks in different cancer types, with potential clinical implications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Takeda, H. et al. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat. Genet. 47, 142–150 (2015).
Rad, R. et al. A conditional piggyBac transposition system for genetic screening in mice identifies oncogenic networks in pancreatic cancer. Nat. Genet. 47, 47–56 (2015).
Moriarity, B.S. & Largaespada, D.A. Sleeping Beauty transposon insertional mutagenesis–based mouse models for cancer gene discovery. Curr. Opin. Genet. Dev. 30, 66–72 (2015).
Mann, M.B. et al. Transposon mutagenesis identifies genetic drivers of BrafV600E melanoma. Nat. Genet. 47, 486–495 (2015).
Jones, K.B. Transposon mutagenesis disentangles osteosarcoma genetic drivers. Nat. Genet. 47, 564–565 (2015).
Vassiliou, G.S. et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 43, 470–475 (2011).
Rad, R. et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 330, 1104–1107 (2010).
Bard-Chapeau, E.A. et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat. Genet. 46, 24–32 (2014).
Pérez-Mancera, P.A. et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486, 266–270 (2012).
Rangel, R. et al. Transposon mutagenesis identifies genes that cooperate with mutant Pten in breast cancer progression. Proc. Natl. Acad. Sci. USA 113, E7749–E7758 (2016).
Ahmad, I. et al. Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer. Proc. Natl. Acad. Sci. USA 113, 8290–8295 (2016).
Dupuy, A.J., Akagi, K., Largaespada, D.A., Copeland, N.G. & Jenkins, N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005).
Luo, G. et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26, 424–429 (2000).
Hollander, M.C., Blumenthal, G.M. & Dennis, P.A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer 11, 289–301 (2011).
Friedrich, M.J. et al. Genome-wide transposon screening and quantitative insertion site sequencing for cancer gene discovery in mice. Nat. Protoc. 12, 289–309 (2017).
de Ridder, J., Uren, A., Kool, J., Reinders, M. & Wessels, L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLOS Comput. Biol. 2, e166 (2006).
Collier, L.S., Carlson, C.M., Ravimohan, S., Dupuy, A.J. & Largaespada, D.A. Cancer gene discovery in solid tumors using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276 (2005).
Karreth, F.A. et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395 (2011).
Keng, V.W. et al. PTEN and NF1 inactivation in Schwann cells produces a severe phenotype in the peripheral nervous system that promotes the development and malignant progression of peripheral nerve sheath tumors. Cancer Res. 72, 3405–3413 (2012).
Dorr, C. et al. Transposon mutagenesis screen identifies potential lung cancer drivers and CUL3 as a tumor suppressor. Mol. Cancer Res. 13, 1238–1247 (2015).
Alimonti, A. et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 42, 454–458 (2010).
Trotman, L.C. et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1, E59 (2003).
Ai, J. et al. Concomitant loss of Eaf2 (U19) and Pten synergistically promotes prostate carcinogenesis in the mouse model. Oncogene 33, 2286–2294 (2014).
Patel, R. et al. Sprouty2, PTEN and PP2A interact to regulate prostate cancer progression. J. Clin. Invest. 123, 1157–1175 (2013).
Fernández-Marcos, P.J. et al. Simultaneous inactivation of Par-4 and Pten in vivo leads to synergistic NF-κB activation and invasive prostate carcinoma. Proc. Natl. Acad. Sci. USA 106, 12962–12967 (2009).
Carver, B.S. et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat. Genet. 41, 619–624 (2009).
Abate-Shen, C. et al. Nkx3.1;Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 63, 3886–3890 (2003).
Di Cristofano, A., De Acetis, M., Koff, A., Cordon-Cardo, C. & Pandolfi, P.P. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat. Genet. 27, 222–224 (2001).
An, O. et al. NCG 4.0: the network of cancer genes in the era of massive mutational screenings of cancer genomes. Database 7, bau015 (2014).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Cooper, C.S. et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 47, 367–372 (2015).
Boutros, P.C. et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet. 47, 736–745 (2015).
Taylor, B.S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010).
Barbieri, C.E. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Grasso, C.S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Ding, L. et al. CBP loss cooperates with PTEN haploinsufficiency to drive prostate cancer: implications for epigenetic therapy. Cancer Res. 74, 2050–2061 (2014).
Chen, J.L. et al. Deregulation of a Hox protein regulatory network spanning prostate cancer initiation and progression. Clin. Cancer Res. 18, 4291–4302 (2012).
Koon, H.B., Ippolito, G.C., Banham, A.H. & Tucker, P.W. FOXP1: a potential therapeutic target in cancer. Expert Opin. Ther. Targets 11, 955–965 (2007).
Takayama, K. et al. Integrative analysis of FOXP1 function reveals a tumor-suppressive effect in prostate cancer. Mol. Endocrinol. 28, 2012–2024 (2014).
Demichelis, F. et al. Distinct genomic aberrations associated with ERG-rearranged prostate cancer. Genes Chromosom. Cancer 48, 366–380 (2009).
Naguib, A. et al. PTEN functions by recruitment to cytoplasmic vesicles. Mol. Cell 58, 255–268 (2015).
Leslie, N.R., Batty, I.H., Maccario, H., Davidson, L. & Downes, C.P. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene 27, 5464–5476 (2008).
Tay, Y. et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357 (2011).
Sarver, A.L. & Subramanian, S. Competing endogenous RNA database. Bioinformation 8, 731–733 (2012).
Carver, B.S. et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586 (2011).
White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 12, 401–410 (2012).
McKnight, N.C. et al. Genome-wide siRNA screen reveals amino acid starvation–induced autophagy requires SCOC and WAC. EMBO J. 31, 1931–1946 (2012).
Joachim, J. et al. Activation of ULK kinase and autophagy by GABARAP trafficking from the centrosome is regulated by WAC and GM130. Mol. Cell 60, 899–913 (2015).
Fernández, A.F. & López-Otín, C. The functional and pathologic relevance of autophagy proteases. J. Clin. Invest. 125, 33–41 (2015).
Yin, Y. & Shen, W.H. PTEN: a new guardian of the genome. Oncogene 27, 5443–5453 (2008).
Milella, M. et al. PTEN: multiple functions in human malignant tumors. Front. Oncol. 5, 24 (2015).
Qu, X. et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 (2003).
Santanam, U. et al. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev. 30, 399–407 (2016).
Skarnes, W.C. et al. A conditional-knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011).
Izsvák, Z. et al. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 277, 34581–34588 (2002).
Lee, E.C. et al. A highly efficient Escherichia coli–based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 (2001).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P.T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Temiz, N.A. et al. RNA sequencing of Sleeping Beauty transposon-induced tumors detects transposon–RNA fusions in forward genetic cancer screens. Genome Res. 26, 119–129 (2016).
Wu, T.D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G.D. Enrichment map: a network-based method for gene set enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Acknowledgements
We thank the staff members of the Research Support Facility at the Wellcome Trust Sanger Institute, the Laboratory of Molecular Medicine at IMOMA, and those at the Transgenic Animal Unit, the Molecular Histopathology Unit, the Department of Biochemistry and Molecular Biology and the Biobank of the Principality of Asturias at IUOPA, for excellent technical assistance. This work was supported by grants from the Wellcome Trust (grant no. 098051; A.B.), the Ministerio de Economía y Competitividad–Spain (grant no. SAF2014-52413; C.L.-O.) and the German Research Society (grant no. SFB1243; R.R.), as well as by funding from the Fundación María Cristina Masaveu Peterson (J.C.), the Fundación Centro Médico de Asturias (J.C.), the Fundación Bancaria Caja de Ahorros de Asturias/Liberbank (J.C. and A.A.), FEBS (J.d.L.R. and J.C.), CIBERONC (Plan Feder) (C.L.-O.), the Progeria Research Foundation (C.L.-O.), the EDP Foundation (C.L.-O.) and the German Cancer Consortium (R.R.). G.S.V. is funded by a Wellcome Trust Senior Fellowship in Clinical Science (WT095663MA). J.d.l.R. is a recipient of a FEBS Long-Term Fellowship and was a recipient of a fellowship from the Fundación María Cristina Masaveu Peterson during part of this work. J.C. was a recipient of a FEBS Long-Term Fellowship in the initial phases of this work.
Author information
Authors and Affiliations
Contributions
J.d.l.R., R.R., C.L.-O., A.B. and J.C. designed the study; J.d.l.R., J.W., R.R. and J.C. generated mouse alleles and cohorts, and performed experiments; J.d.l.R., J.W., L.R., Q.L., M.A.L., G.S.V., R.R. and J.C. performed mouse necropsies; A.A., M.S.F.-G., M.T.F.-G. and G.J.H. performed histopathological analysis; J.d.l.R., M.J.F., Y.L and H.P. did bioinformatics analyses; J.d.l.R., C.L.-O. and J.C. interpreted results; M.J.F., S.B.d.Q., I.N., E.M., A.S. and R.F. contributed to some of the experiments; R.R., C.L.-O., A.B. and J.C supervised the study; and J.d.l.R. and J.C. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 (PDF 10687 kb)
Supplementary Tables
Supplementary Tables 1–36 (XLSX 18564 kb)
Rights and permissions
About this article
Cite this article
de la Rosa, J., Weber, J., Friedrich, M. et al. A single-copy Sleeping Beauty transposon mutagenesis screen identifies new PTEN-cooperating tumor suppressor genes. Nat Genet 49, 730–741 (2017). https://doi.org/10.1038/ng.3817
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3817
This article is cited by
-
Genetic alterations of the SUMO isopeptidase SENP6 drive lymphomagenesis and genetic instability in diffuse large B-cell lymphoma
Nature Communications (2022)
-
Single allele loss-of-function mutations select and sculpt conditional cooperative networks in breast cancer
Nature Communications (2021)
-
A resource of targeted mutant mouse lines for 5,061 genes
Nature Genetics (2021)
-
CRISPR and transposon in vivo screens for cancer drivers and therapeutic targets
Genome Biology (2020)
-
PiggyBac mutagenesis and exome sequencing identify genetic driver landscapes and potential therapeutic targets of EGFR-mutant gliomas
Genome Biology (2020)