Abstract
Neurodevelopmental disorders with periventricular nodular heterotopia (PNH) are etiologically heterogeneous, and their genetic causes remain in many cases unknown. Here we show that missense mutations in NEDD4L mapping to the HECT domain of the encoded E3 ubiquitin ligase lead to PNH associated with toe syndactyly, cleft palate and neurodevelopmental delay. Cellular and expression data showed sensitivity of PNH-associated mutants to proteasome degradation. Moreover, an in utero electroporation approach showed that PNH-related mutants and excess wild-type NEDD4L affect neurogenesis, neuronal positioning and terminal translocation. Further investigations, including rapamycin-based experiments, found differential deregulation of pathways involved. Excess wild-type NEDD4L leads to disruption of Dab1 and mTORC1 pathways, while PNH-related mutations are associated with deregulation of mTORC1 and AKT activities. Altogether, these data provide insights into the critical role of NEDD4L in the regulation of mTOR pathways and their contributions in cortical development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
Primary accessions
NCBI Reference Sequence
Referenced accessions
NCBI Reference Sequence
Protein Data Bank
References
Caviness, V.S. Jr., Takahashi, T. & Nowakowski, R.S. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 18, 379–383 (1995).
Rakic, P. & Caviness, V.S. Jr. Cortical development: view from neurological mutants two decades later. Neuron 14, 1101–1104 (1995).
Barkovich, A.J., Guerrini, R., Kuzniecky, R.I., Jackson, G.D. & Dobyns, W.B. A developmental and genetic classification for malformations of cortical development: update 2012. Brain 135, 1348–1369 (2012).
Francis, F. et al. Human disorders of cortical development: from past to present. Eur. J. Neurosci. 23, 877–893 (2006).
Guerrini, R. & Dobyns, W.B. Malformations of cortical development: clinical features and genetic causes. Lancet Neurol. 13, 710–726 (2014).
Fox, J.W. et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21, 1315–1325 (1998).
Parrini, E. et al. Periventricular heterotopia: phenotypic heterogeneity and correlation with filamin A mutations. Brain 129, 1892–1906 (2006).
Ferland, R.J. et al. Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Hum. Mol. Genet. 18, 497–516 (2009).
Carabalona, A. et al. A glial origin for periventricular nodular heterotopia caused by impaired expression of filamin-A. Hum. Mol. Genet. 21, 1004–1017 (2012).
Lian, G. et al. Filamin A regulates neural progenitor proliferation and cortical size through Wee1-dependent Cdk1 phosphorylation. J. Neurosci. 32, 7672–7684 (2012).
Sheen, V.L. et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat. Genet. 36, 69–76 (2004).
Conti, V. et al. Periventricular heterotopia in 6q terminal deletion syndrome: role of the C6orf70 gene. Brain 136, 3378–3394 (2013).
Poirier, K. et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 45, 639–647 (2013).
Mirzaa, G.M. et al. Characterisation of mutations of the phosphoinositide-3-kinase regulatory subunit, PIK3R2, in perisylvian polymicrogyria: a next-generation sequencing study. Lancet Neurol. 14, 1182–1195 (2015).
Kumar, S., Tomooka, Y. & Noda, M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 185, 1155–1161 (1992).
Sudol, M., Chen, H.I., Bougeret, C., Einbond, A. & Bork, P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 369, 67–71 (1995).
Rizo, J. & Südhof, T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273, 15879–15882 (1998).
Huang, L. et al. Structure of an E6AP–UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science 286, 1321–1326 (1999).
Goel, P., Manning, J.A. & Kumar, S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene 557, 1–10 (2015).
Garrone, N.F., Blazer-Yost, B.L., Weiss, R.B., Lalouel, J.M. & Rohrwasser, A. A human polymorphism affects NEDD4L subcellular targeting by leading to two isoforms that contain or lack a C2 domain. BMC Cell Biol. 10, 26 (2009).
Hsia, H.E. et al. Ubiquitin E3 ligase Nedd4-1 acts as a downstream target of PI3K/PTEN–mTORC1 signaling to promote neurite growth. Proc. Natl. Acad. Sci. USA 111, 13205–13210 (2014).
Franco, S.J., Martinez-Garay, I., Gil-Sanz, C., Harkins-Perry, S.R. & Müller, U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron 69, 482–497 (2011).
Olson, E.C., Kim, S. & Walsh, C.A. Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression. J. Neurosci. 26, 1767–1775 (2006).
Sekine, K., Honda, T., Kawauchi, T., Kubo, K. & Nakajima, K. The outermost region of the developing cortical plate is crucial for both the switch of the radial migration mode and the Dab1-dependent “inside-out” lamination in the neocortex. J. Neurosci. 31, 9426–9439 (2011).
Sekine, K. et al. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron 76, 353–369 (2012).
Lifton, R.P., Gharavi, A.G. & Geller, D.S. Molecular mechanisms of human hypertension. Cell 104, 545–556 (2001).
Boase, N.A. & Kumar, S. NEDD4: the founding member of a family of ubiquitin–protein ligases. Gene 557, 113–122 (2015).
Moon, U.Y. et al. Impaired reelin-Dab1 signaling contributes to neuronal migration deficits of tuberous sclerosis complex. Cell Rep. 12, 965–978 (2015).
Gao, S. et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell 36, 457–468 (2009).
Yu, J.S. et al. PI3K/mTORC2 regulates TGF-β/activin signalling by modulating Smad2/3 activity via linker phosphorylation. Nat. Commun. 6, 7212 (2015).
Wiesner, S. et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 (2007).
Bruce, M.C. et al. Regulation of Nedd4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem. J. 415, 155–163 (2008).
Wang, J. et al. Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain–mediated auto-inhibition. J. Biol. Chem. 285, 12279–12288 (2010).
Escobedo, A. et al. Structural basis of the activation and degradation mechanisms of the E3 ubiquitin ligase Nedd4L. Structure 22, 1446–1457 (2014).
Honda, T. & Nakajima, K. Proper level of cytosolic disabled-1, which is regulated by dual nuclear translocation pathways, is important for cortical neuronal migration. Cereb. Cortex 26, 3219–3236 (2016).
Rivière, J.B. et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 44, 934–940 (2012).
Jansen, L.A. et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 138, 1613–1628 (2015).
Wright, C.F. et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 385, 1305–1314 (2015).
Cau, E., Gradwohl, G., Fode, C. & Guillemot, F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development 124, 1611–1621 (1997).
Kielar, M. et al. Mutations in Eml1 lead to ectopic progenitors and neuronal heterotopia in mouse and human. Nat. Neurosci. 17, 923–933 (2014).
Acknowledgements
We are grateful to the patients and their families for their participation. We thank D. Rotin and C. Jiang (The Hospital for Sick Children) for kindly providing humanV5-tagged Nedd4-2 constructs (pcDNA3.1-nV5 wild type, CS and Y971A constructs) and L. Lindner, M. Ruff, M. Macias and F. Francis for their thoughtful comments and help. We thank investigators from the Epi4K Consortium and the Epilepsy Phenome/Genome Project for contributing NEDD4L-related genotype and phenotype data. This work was supported by funding from Strasbourg University and grant ANR-10-LABX-0030-INRT, a French State Fund managed by the Agence Nationale de la Recherche under the frame program Investissements d'Avenir ANR-10-IDEX-0002-02, the Fondation pour la Recherche Médicale (FRM funding within the framework of the program Equipe FRM; J.C–DEQ20130326477), the Fondation Maladies Rares, the Fondation NRJ–Institut de France, Agence National de Recherche (ANR Blanc 1103 01, project R11039KK; ANR E-Rare-012-01, project E10107KP; ANR-13-BSV-0009-01) and European Union FP7 project GENECODYS (grant 241995) and DESIRE (grant agreement 602531), and funding provided from the National Institute of Neurological Disorders and Stroke to the Epi4k Consortium and the Epilepsy Phenome/Genome Project (NS053998, NS077364, NS077274, NS077303 and NS077276). This study was also supported in part by the NIHR Biomedical Research Centre Oxford with funding from the UK Department of Health's NIHR Biomedical Research Centre funding scheme. The views expressed in this publication are those of the authors and not necessarily those of the UK Department of Health.
Author information
Authors and Affiliations
Consortia
Contributions
L.B., H.J., E.L.I. and M.-V.H. conceived and designed the experiments, performed the experiments, performed statistical analysis and analyzed the data related to cellular, IUEP and functional studies. S.S. provided technical assistance and performed ubiquitination experiments. N.D. provided technical assistance, performed expression and genetic studies, and prepared reagents. J.C.-S., K.A.M., B.I., U.W.L., A.P., N.B.W., R.G., D.K., C.B., the DDD study, U.K. and N.B.-B. contributed clinical and imaging data and follow-up of patients and families. P.T. and G.M. provided assistance for IUEP studies. A.T.P., J.C.T., K.P., Y.S., N.L., G.R., B.S., A.E. and R.T. contributed to genetic studies and analysis of variants in candidate genes and screened DNA from subjects. M.M. and J.G. performed expression studies during brain development. L.N. and J.G. contributed reagents and material, as well as critical suggestions for functional studies. T.S., the DDD study, A.B., R.O., C.M., P.N. and J.-F.D. contributed to the generation of whole-exome sequencing, bioinformatics tools and analysis of sequencing data. I.S. conceived and designed ubiquitination experiments. J.C. conceived, coordinated and supervised the study, designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members and affiliations appears at http://dx.doi.org/10.1101/049056.
Integrated supplementary information
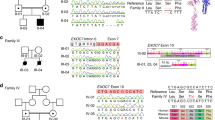
Supplementary Figure 1 Pedigrees and Sanger sequencing traces of NEDD4L mutations.
(a–f) Chromatograms for family members showing normal and mutated sequences of NEDD4L. For family P347, note the mother’s chromatogram showing a small peak corresponding to the mutated allele and suggesting somatic mosaicism. For family DDDP110533, the Sanger sequence trace for the patient and the IGV screenshot confirm the mutation and show the high sequencing coverage (>70×) in the trio and the absence of the mutation in the sequencing reads of the parents. None of these validated variants was reported in the available genomic databases, and to strengthen their relevance we reviewed trio data sets corresponding to the 3,287 individuals with developmental disorders highlighted in the recent report of the Deciphering Developmental Disorders study (http://dx.doi.org/10.1101/049056). Except the de novo mutation located in the N-terminal part of the protein, p.Pro294Arg, in a patient with infantile spasm (IS) and without PNH (Allen, 2013 #91), no other de novo mutation affecting the HECT domain was reported. These data are in line with the fact that the NEDD4L HECT domain is the most intolerant to mutations across the gene as assessed by subRVIS (http://www.subrvis.org/).
Supplementary Figure 2 Illustration of the structure of the NEDD4L HECT domain and the position of PNH-associated alterations.
(a) Multiple-sequence alignment showing the high conservation of the HECT domain regions containing the Tyr679, Gln694, Glu893 and Arg897 residues. (b,c,e,g,i) Representation (visualized with Chimera) of the N-lobe and C-lobe of the HECT domain (PDB 2ONI) that are shown in gray and magenta, respectively, the flexible hinge in yellow, the cysteine residue of the catalytic site in red, the PY motif in mint and the position of the different changed residues. (d,f,h,j) Models of the HECT domain constructed with the Phyre web portal and based on PDB model 2ONI and the sequence of the HECT domain from the human NEDD4L protein Q96PU5 with altered residues Cys679, His694, Lys893 and Gln897. Inspection of the protein structure shows that the Glu893 and Arg897 residues are located on the surface of the α3 helix of the C-lobe that contains the active-site cysteine that forms the thioester with ubiquitin, the Tyr679 residue is located between the α2 helix and β2 sheet of the N-lobe, and the Gln694 residue is located within the β3 sheet of the N-lobe. Because of the physicochemical property changes (i.e., from acidic to basic for p.Glu893Lys or from basic to uncharged for p.Arg897Gln), NEDD4L function could be disrupted through the effects of the mutations on stability, interference with functional properties of the HECT domain or deregulation of interactions with substrates.
Supplementary Figure 3 NEDD4L expression in mouse developing cortex.
(a) In situ hybridization with Nedd4l probe of an E15 brain section showing cortical expression and distribution of Nedd4l. (b) qRT–PCR analyses showing cortical expression of Nedd4L transcripts at E12.5, E14.5, E16.5 and E18.5. Scale bar, 200 μm.
Supplementary Figure 4 Expression and cellular localization of wild-type and NEDD4L mutants in cultured neuronal cells.
Wild-type and mutant NEDD4L cDNA constructs were transfected into primary cultures of neuronal cells, and NEDD4L expression was analyzed by immunofluorescence labeling. Immunostaining (green) shows a cytoplasmic distribution, with enrichment in the periphery of neuronal cells transfected with the wild-type NEDD4L construct, whereas the p.Gln694His, p.Glu893Lys and p.Arg897Gln mutants are not detectable.
Supplementary Figure 5 Expression of wild-type and NEDD4L control variants and analysis of their consequences on mTORC1 and AKT activities.
(a) Tomato/DAPI and NEDD4L detection in N2A cells transfected with empty vector, wild-type and NEDD4L control variant cDNA constructs. NEDD4L immunostaining (green) shows similar cytoplasmic distribution, with enrichment in the cellular of N2A cells, of wild-type and NEDD4L control variants. (b) Immunoblot analyses showing similar expression levels and phosphorylation patterns of S6 and AKT resulting from transfection of N2A cells with wild-type NEDD4L and the three control variants reported in the ExAC database (c.698C>T, S233L; c.535T>A, S179T and c.2614G>A, G872S (located in the HECT domain)).
Supplementary Figure 6 qRT–PCR analysis of NEDD4L transcripts in N2A cells transfected with wild-type and mutant NEDD4L cDNA constructs.
Real-time qPCRs were performed from transfected N2A cells using the SYBR Green method on total, DNase-treated RNA samples, using NEDD4L-specific primers. GFP was used for normalization.
Supplementary Figure 7 Analysis of NEDD4L mutant (p.Arg897Gln) ubiquitination activity.
(a) Immunoprecipitation against V5-tagged Nedd4l and analysis by immunoblotting using antibodies to ubiquitin, V5 and NEDD4L. We can observe in the immunoprecipitate (IP) a band corresponding to NEDD4L and a smear of the ubiquitinated protein. (b) Wild-type and mutant NEDD4L were immunopurified from transfected N2A cell lysates and incubated with ATP, E1 enzyme and E2 (UbcH7) enzyme with (+) or without (–) ubiquitin. Reaction mixtures were analyzed by immunoblotting using antibodies to V5 and NEDD4L. Note that, despite the unbalanced amounts of wild-type and NEDD4L mutant resulting from the instability of mutant NEDD4L, in the presence of ubiquitin both wild-type and mutant NEDD4L exhibit enhanced ubiquitination activity, as illustrated by the high-molecular-mass smear representing self-ubiquitinated NEDD4L.
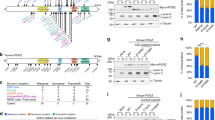
Supplementary Figure 8 In utero knockdown of mouse Nedd4l expression by RNAi and analysis of neuronal positioning in the isocortex.
(a) The images show coronal sections of mouse brains 4 d after electroporation at E14.5 with the Tomato-encoding reporter construct either in combination with the shRNA targeting Nedd4l mRNA or the corresponding scrambled controls (scrambled-sh). Scale bar, 100 μm. (b) Fluorescent neuron positioning was quantified in three regions: VZ/SVZ, IZ and CP. Bars represent the means of fluorescent neurons ± s.e.m. of independent brains (Nedd4l-sh, n = 4; scrambled-sh, n = 5). Note the absence of significant differences between the distribution in the three regions of neuronal cells electroporated with shRNA and the control scrambled-sh.
Supplementary Figure 9 Effect of wild-type and NEDD4L mutants on neuronal positioning, proliferation and differentiation.
(a) The images show coronal sections of mouse brains at P2 and electroporated at E14.5 with either empty vector, wild-type NEDD4L or mutant NEDD4L cDNA constructs in combination with an RFP-encoding vector (Tomato). Sections were stained for a neuronal marker, NeuN (magenta), and an upper-layer maker, Cux1 (green), and were counterstained with DAPI (blue) to determine neuronal positioning in the white matter (WM), layers VI, V and II–IV. Scale bar, 100 μm. (b) Fluorescent neurons were quantified in the P2 brain regions highlighted in a: WM: empty vector (EV) versus WT, P = 0.0004, EV versus p.Glu893Lys, P = 0.0078; VI: EV versus WT, P = 0.0091; V: EV versus p.Arg897Gln, P = 0.0185; II–IV: EV versus WT, P < 0.0001, EV versus p.Glu893Lys, P = 0.0024, EV versus p.Arg897Gln, P = 0.0448). Bars represent the means of fluorescent neurons ± s.e.m (EV, n = 4; WT, n = 3; p.Glu893Lys, n = 4; p.Arg897Gln, n = 3) in the white matter, layers VI, V and IV–II, which were determined by examination of DAPI-stained coronal sections immunostained with NeuN and Cux1 markers shown in a. (c) Immunofluorescence staining of NEDD4L (magenta) in electroporated cells (green) in the IZ of P2 brains electroporated at E14.5 with each construct. Scale bar, 10 μm. (d) Representative image of brain slices at E16.5 immunostained for the mitotic marker PH3 (magenta) used for mitotic index quantification in the VZ. (e,f) Representative images of brain slices at E16.5 immunostained for a radial glial cell marker, Pax6 (magenta), an intermediate progenitor marker, Tbr2 (yellow), and a proliferating progenitor marker (Ki67; magenta) used for quantification of Pax6/Tbr2 pools in the VZ/SVZ (see Results section) and proliferating progenitors. Doted lines indicate the VZ and SVZ. (g) Quantification of proliferative cells in the VZ and SVZ. Bars represent the means of double-positive Ki67+Tomato+ ± s.e.m of analyzed independent brains in the VZ and SVZ (empty vector, n = 3; WT, n = 3; p.Arg897Gln, n = 3; p.Glu893Lys, n = 3). Scale bars in f, 50 μm. (h) Cortical slices at E18.5 stained with an antibody against Cux1 (magenta) showed that misplaced neurons electroporated (green) with wild-type and PNH-related mutations in deep layers of the cortex ectopically expressed the upper-layer marker. Right panels are enlargements of the regions highlighted by white boxes in the IZ. Scale bar, 100 μm (left) and 10 μm (right).
Supplementary Figure 10 Effect of wild-type and NEDD4L mutants on neuronal terminal translocation.
(a) Representative images of brain sections showing electroporated neurons with Tomato reporter and NEDD4L constructs at E14.5 analyzed at P2 for the Cux1 (yellow) and NeuN (magenta) markers. (b) Quantification of electroporated neurons in the upper layers of corticies reflecting analysis of the terminal translocation process. Expression of wild-type and NEDD4L mutants leads to enrichment in the down Cux1 region (empty vector versus WT, P < 0.0001, empty vector versus p.Glu893Lys, P = 0.0004, empty vector versus p.Arg897Gln, P = 0.0003), while PCZ is significantly depleted (empty vector versus WT, P < 0.0001, empty vector versus p.Glu893Lys, P < 0.0001, empty vector versus p.Arg897Gln, P = 0.0005), Scale bar, 50 μm. Bars represent the means of electroporated neurons ± s.e.m. of analyzed independent brains (empty vector, n = 3; WT, n = 3; p.Glu893Lys, n = 3; p.Arg897Gln, n = 3).
Supplementary Figure 11 Deregulation of AKT and Smad2/3 activation resulting from PNH-related NEDD4L mutant.
Representative immunoblots showing the effect of expression of wild-type and mutant NEDD4L constructs on Akt and Smad2/3 phosphorylation under basal condition and upon treatment of cells for 1 h with activin A (AA), an activator of the TGF-β pathway.
Supplementary Figure 12 NEDD4L expression after rapamycin treatment.
Immunofluorescence staining of NEDD4L (magenta) in Tomato-electroporated cells (green) of E18.5 brains electroporated with wild-type NEDD4L cDNA construct at E14.5 and treated daily with rapamycin. Right panels are enlargements of the regions highlighted by white boxes. Scale bars, 100 μm (left) and 10 μm (right). Note that wild-type NEDD4L is still highly expressed in the electroporated cells of rapamycin-treated brains.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Table 1 and Supplementary Note. (PDF 2359 kb)
Supplementary Table 2
Statistical analysis details. (XLSX 45 kb)
Rights and permissions
About this article
Cite this article
Broix, L., Jagline, H., L Ivanova, E. et al. Mutations in the HECT domain of NEDD4L lead to AKT–mTOR pathway deregulation and cause periventricular nodular heterotopia. Nat Genet 48, 1349–1358 (2016). https://doi.org/10.1038/ng.3676
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3676
This article is cited by
-
Periventricular nodular heterotopias is associated with mutation at the FLNA locus-a case history and a literature review
BMC Pediatrics (2023)
-
Genome-wide study of longitudinal brain imaging measures of multiple sclerosis progression across six clinical trials
Scientific Reports (2023)
-
A novel de novo KCNB1 variant altering channel characteristics in a patient with periventricular heterotopia, abnormal corpus callosum, and mild seizure outcome
Journal of Human Genetics (2023)
-
BioE3 identifies specific substrates of ubiquitin E3 ligases
Nature Communications (2023)
-
Downregulation of NEDD4L by EGFR signaling promotes the development of lung adenocarcinoma
Journal of Translational Medicine (2022)