Abstract
Whipworms are common soil-transmitted helminths that cause debilitating chronic infections in man. These nematodes are only distantly related to Caenorhabditis elegans and have evolved to occupy an unusual niche, tunneling through epithelial cells of the large intestine. We report here the whole-genome sequences of the human-infective Trichuris trichiura and the mouse laboratory model Trichuris muris. On the basis of whole-transcriptome analyses, we identify many genes that are expressed in a sex- or life stage–specific manner and characterize the transcriptional landscape of a morphological region with unique biological adaptations, namely, bacillary band and stichosome, found only in whipworms and related parasites. Using RNA sequencing data from whipworm-infected mice, we describe the regulated T helper 1 (TH1)-like immune response of the chronically infected cecum in unprecedented detail. In silico screening identified numerous new potential drug targets against trichuriasis. Together, these genomes and associated functional data elucidate key aspects of the molecular host-parasite interactions that define chronic whipworm infection.
Similar content being viewed by others
Main
Whipworms (T. trichiura) infect an estimated 700 million people worldwide1. They are one of three major groups of soil-transmitted helminths (STHs), the others being Ascaris and hookworms, that impede the socioeconomic development of entire populations. Trichuris parasites invade the epithelium of the colon of their hosts, particularly the cecum, where they may persist for years and can cause colitis, anemia and Trichuris dysentery syndrome. T. muris is an established laboratory mouse model for human trichuriasis and shares with T. trichiura many aspects of its biology, including the specialized niche within the host. Work on T. muris has been pivotal in defining the crucial role of helper T cell 2 (TH2) immune responses and interleukin (IL)-13 in protective immunity to parasitic helminths2, and deliberate infection with whipworm eggs, typically of the porcine parasite Trichuris suis, is emerging as a novel therapy option for patients with immune-mediated diseases3,4. Whipworms have a simple and direct life cycle, and, unlike the related parasite Trichinella spiralis, whipworm larvae do not form cysts in muscle tissue but reside exclusively in the intestine. Bacillary cells and stichocytes are distinctive cell types found only in clade I nematodes5 and are located in the slender anterior part of the adult whipworm that is burrowed within the intestinal epithelium. The bulbous posterior end of the whipworm lies free in the intestinal lumen and harbors the reproductive organs, giving adult Trichuris parasites their characteristic whip-like morphology.
Here we present high-quality genome sequences for T. trichiura and T. muris, the first duo of a major human STH and its mouse counterpart. We resolve chromosomal sequences and infer sex chromosome–specific parasite genes and new potential drug targets. On the basis of high-throughput transcriptomics data, we identify whipworm proteins that are highly expressed in the anterior region of the parasite, that is, in intimate contact with the cytoplasm of host intestinal cells and the immune system. Gene expression data from mice with low-dose whipworm infection provide a detailed description of a regulated TH1-like immune response to the infected cecum that is not limited to the immediate site of infection. The availability of these two important whipworm genomes and the integration of parasite and host data presented here will underpin future efforts to control these parasites and exploit their immunological interplay for human benefit.
Results
Genome structure and content
We produced a 75.2-Mb high-quality draft genome assembly from a clinically isolated adult male T. trichiura using Illumina technology. In parallel, multiple technologies and manual finishing were used to produce an 85-Mb reference genome assembly from the more readily available mouse parasite species T. muris (Online Methods and Supplementary Note). More than half of the T. muris and T. trichiura genomes assembled into scaffolds of at least 1,580 kb and 71 kb, respectively (Table 1). Despite their differences in contiguity, both genomes were determined to be 94.8% complete using the Core Eukaryotic Genes Mapping Approach (CEGMA)6. The T. muris genome comprises two pairs of autosomes and one pair of X and Y sex chromosomes (2n = 6)7, whereas the karyotype of T. trichiura has so far not been reported. In Trichinella species of the same phylogenetic clade, the XO sex-determination system is used with 2n = 6 for female (XX) and 2n = 5 for male (X0) worms8. To explore the architecture of the genome, assembly scaffolds for T. muris were joined into superscaffolds by optical mapping (assembly version 4.0; Table 1). Clustering of one-to-one ortholog patterns between T. muris and T. spiralis identified three distinct linkage groups in each species, providing strong evidence that chromosome-level synteny has been preserved across genera (Fig. 1a). Contrasting with the observed macrosynteny, small-scale gene order has mostly been lost among the three species (Fig. 1a and Supplementary Fig. 1). Such extensive intrachromosomal rearrangements have also been found in other nematode lineages9,10,11,12 and appear to be a hallmark of nematode genome evolution.
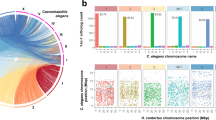
(a) Mapping one-to-one gene orthologs (as determined by Inparanoid) between genome scaffolds for T. spiralis and T. muris (genome assembly v4) followed by clustering of the resulting ortholog pattern identifies three large linkage groups in each genome (left; Supplementary Note). The table lists the number of one-to-one gene orthologs for individual comparisons between the 11 longest scaffolds of T. spiralis and the 17 longest scaffolds of T. muris, which represent 85.2% and 89.0% of the respective genomes. The relationship of one-to-one gene orthologs shows high-level and cross-genus synteny between T. muris and T. spiralis (right). The linkage group shown in yellow is putatively identified as the sex-specific X chromosome. (b) Median relative coverage of high-throughput sequencing reads was derived for a pool of 11 female T. muris parasites or single male parasites (T. muris and T. trichiura) and was calculated per 10-kb window across all genome scaffolds that were assigned to 1 of the 3 linkage groups. In females, mapped sequence read coverage is even across all three linkage groups, whereas, in males, read coverage exhibits a bimodal distribution. In particular, the linkage group to which scaffolds belong separates well with either of the two peaks of relative read coverage. Scaffolds of linkage group X are associated with half the median read coverage found for scaffolds of linkage groups 1 and 2. (c) Levels of heterozygosity correlate strongly with affiliation with one of the three linkage groups. Both the 0.5-fold relative read coverage and the very low apparent heterozygosity of linkage group X are consistent with the corresponding scaffolds representing the sex-specific X chromosome, which is expected to occur in a single copy in the diploid genome of a male Trichuris parasite. In box plots, the center line is the median, the box shows 25% to 75% quantiles (interquartile range), the top whisker represents the highest data point below (75% quartile + 1.5 × interquartile range) and the bottom whisker shows the lowest data point above (25% quartile + 1.5 × interquartile range).
To assign genomic scaffolds to chromosomal locations, we used mapped resequencing data from females (T. muris only) and single males (T. muris and T. trichiura). From the males, one linkage group could be putatively identified as the X chromosome, on the basis of an almost complete lack of heterozygosity and half the relative sequence coverage compared with the other two linkage groups that are presumed to be pairs of autosomes (Fig. 1b,c, Supplementary Fig. 2a–c and Supplementary Table 1). In contrast, the putative X-chromosomal scaffolds from T. muris females showed equivalent read coverage and a similar level of heterozygosity to the putative autosomes. Furthermore, the putative X-chromosomal scaffolds were significantly (P < 1 × 10−12) enriched for genes that were transcriptionally upregulated in female worms (Supplementary Table 2a).
From differential sequence coverage in males and females (Online Methods, Supplementary Fig. 2e,f, Supplementary Table 1c and Supplementary Note), putative T. muris Y chromosome sequences were identified, equivalent to 24.4 Mb of sequence when the estimated true copy numbers of the large number of collapsed repeats were taken into account (Supplementary Table 1d). Despite the similar estimated sizes for the T. muris sex chromosomes, the X chromosome (26.9 Mb) contained 2,137 genes, whereas the Y chromosome had non-redundant gene content corresponding to fewer than 70 genes, of which most were of unknown function or were transposon related (Supplementary Table 2b). We further identified putative centromeric repeat sequences on three T. muris scaffolds that together were estimated to constitute 5.33 Mb (Supplementary Fig. 3, Supplementary Table 1d and Supplementary Note).
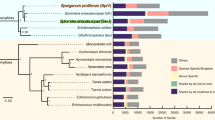
In T. trichiura, 9,650 genes were predicted, and in T. muris, aided by deep transcriptome sequencing, 11,004 genes were found (Table 1). Predicted proteins from both species clustered together with sequences from other nematodes and outgroups in a total of 7,354 gene families. Of these, 5,868 families contained genes from both species, that is, 6,629 and 6,641 genes from T. muris and T. trichiura, respectively. Thus, the majority of Trichuris genes are orthologs shared by both species (Fig. 2a,b). Despite the widespread loss of gene collinearity, the usefulness of the mouse model is emphasized by the highly similar proteomes of the two species—5,060 one-to-one protein orthologs exhibited an average amino acid identity of 79%. In this comparison, 2,350 T. trichiura and 3,817 T. muris genes appeared to be species specific and were particularly enriched in hypothetical proteins of no known function (for example, 76% of the T. muris species-specific genes compared to 35% across the genome). Of the total 3,953 and 2,014 hypothetical proteins encoded by T. muris and T. trichiura, respectively, 434 (11.0%) and 253 (12.6%) sequences were predicted by Phobius to contain signal peptides, and 517 (13.1%) and 359 (17.8%) sequences were predicted to contain transmembrane domains. For those sequences that had functional annotation, Gene Ontology (GO) term enrichment analysis suggested that extracellular proteins, proteases and protease inhibitors were particularly abundant among the species-specific proteins in T. muris. One large gene family of unknown function in T. muris consisted of a lineage-specific expansion of over 40 T. muris genes and 6 T. trichiura genes, with only 1 or 2 copies found in other nematode species (FAM217; Supplementary Table 3 and Supplementary Data Set 1).
(a) Phylogenetic analysis of genome content. The tree shown is a maximum-likelihood phylogeny based on a concatenated alignment of single-copy orthologs. Values on edges represent the inferred numbers of births (+) and deaths (−) of gene families along that edge. The scale bar shows expected number of amino acid substitutions per residue. Pie charts represent the gene family composition of each genome: the area of the circle is proportional to the predicted proteome size, and wedges represent the numbers of proteins predicted to be either singletons (not members of any gene family; yellow), members of families common to the eight genomes (red), members of gene families present only in a single genome (blue) and members of all other gene families (green). (b) Euler diagrams of shared presence and absence of gene families in clade I nematodes and the model nematode C. elegans. Note that the approach in a cannot distinguish the polarity of changes at the base of the tree; thus, for example, the value of 262 gene family gains on the basal branch will include gene families lost on the branch leading to Homo sapiens.
Patterns of parasite gene expression
To better understand the parasite's unusual niche within the mucosa of the large intestine, we generated and compared transcriptome-wide gene expression data for several biological samples of T. muris, then performing protein domain and GO term enrichment analyses. The stichosome and bacillary band in the whipworm anterior region have been suggested to be involved in nutrient uptake, digestion and host-parasite interactions, for which they would be perfectly placed morphologically. Nevertheless, their exact biological roles are still poorly understood. Our data showed that transcripts in the T. muris anterior region were dominated by chymotrypsin A–like serine proteases and by protease inhibitors with similarity to secretory leukocyte peptidase inhibitor (SLPI) (families S1A and I17, respectively, in the MEROPS peptidase database; Supplementary Fig. 4 and Supplementary Tables 4 and 5). Chymotrypsin A–like serine proteases were encoded by over 75 genes in each of the Trichuris genomes, at numbers far higher than in other nematodes, making them the single largest group of proteases (Supplementary Table 4). In T. muris, three-quarters of these genes (63) were transcriptionally upregulated in the anterior region, and, of these, two-thirds (42) are likely to encode secreted proteins (Supplementary Fig. 4 and Supplementary Tables 6 and 7). These proteases might, therefore, serve digestive or host-immunomodulatory functions or degrade host intestinal mucins that act as a physical barrier to the parasite13,14. In Trichuris, serine proteases are thus more abundant, both in terms of gene number and gene expression, than in other protease families (in particular, cysteine proteases), which is in contrast to the situation in many other nematodes15,16,17 (Supplementary Tables 4 and 5).
Eighty of the 111 protease inhibitors found in T. muris encoded serine protease inhibitors (MEROPS families I1, I2, I15 and I17), with SLPI-like proteins (proteins containing mostly WAP domains) collectively being by far the most abundant protease inhibitor transcripts in the anterior region (Fig. 3a). WAP domains are found in a number of proteins that also include the mammalian whey acidic protein (WAP) and elafins. The T. muris genome contained 44 genes encoding between 1 and 9 WAP domains, whereas T. trichiura and T. spiralis featured 20 and 23 genes encoding such proteins, respectively. Surprisingly, all three trichocephalid species each encoded only a single WAP domain–containing protein (TMUE_s0077003100, TTRE_0000351901 and EFV57447) in which the WAP domain contained the canonical eight cysteine residues that have given this domain its alternative name, 4-disulphide core. These proteins are large and bear some similarity to the mesocentin protein of C. elegans, which exhibits RNA interference (RNAi) phenotypes, such as neuroanatomical defects affecting chemosensory neurons and axons18. In contrast, all other WAP domains in Trichuris and Trichinella had an apparently novel trichocephalid adaptation of exactly six cysteine residues (Fig. 3b and Supplementary Fig. 5). How this modification affects the function of WAP domain–containing proteins is unknown. In addition to their protease inhibitor activity, both mammalian SLPIs and elafins have immunomodulatory, anti-inflammatory and antimicrobial properties, with a role in innate immune defense and wound healing19,20,21. Epithelial cells frequently produce these proteins in response to inflammation to modulate the inflammation process, cytokine secretion and cell recruitment and to favor a TH1-type immune response19. The fact that there are apparently no SLPI-like proteins present in other parasitic helminth lineages, that the corresponding genes are strongly and specifically expressed in the whipworm anterior region and that the majority of SLPI-like proteins are likely secreted suggests that this family carries out nematode clade I–specific functions that may include the inhibition of inflammation in host intestinal epithelial tissue as it is being invaded and wounded by the parasite.
(a) Normalized transcript levels of the 44 genes encoding WAP domain–containing proteins in T. muris, comparing the parasite anterior region with the posterior regions of adult female (F) and male (M) parasites. Indication of significant transcriptional upregulation in a particular pairwise comparison (Up) refers to false discovery rate (FDR) % 0.01 and FDR % 1 × 10−5 when denoted by one asterisk and to FDR % 1 × 10−5 when denoted by two asterisks. SP, signal peptide; WAP, whey acidic protein (Interpro, IPR008197); WR1, cysteine-rich repeat (IPR006150); TIL, trypsin inhibitor–like (IPR002919). For a full version of this figure, see Supplementary Figure 4a. (b) Sequence logos show the conserved and distinct sequence characteristics of the WAP domains (Interpro, IPR008197) found in proteins from H. sapiens, T. trichiura and T. muris. The four canonical disulfide bonds formed by eight cysteine residues are highlighted at the top of the sequence logo for human WAP domains. The sequence logos representing the different species are aligned around the central CXXDXXC motif (where X is any amino acid). For a full version of this figure, see Supplementary Figure 5.
Transcripts for DNase II–like proteins were also particularly highly expressed in the whipworm anterior region (Fig. 4a). T. spiralis encodes 166 DNase II–like proteins22,23 that include the abundant excretory-secretory protein gp43 (ref. 24). T. muris and T. spiralis each encoded one ortholog of the metazoan DNase II (Fig. 4b and Supplementary Fig. 6), which in T. muris exhibited low and uniform transcript levels across the biological samples analyzed. In contrast, all other trichocephalid DNase II–like proteins were so divergent that they were positioned outside the major, well-supported cluster in a phylogenetic tree (Fig. 4b). As in other animals25, the three DNase II–like nucleases in C. elegans (NUC-1, CRN-6 and CRN-7) are involved in apoptosis and development26. A homologous protein in starfish (plancitoxin) has been reported to be able to enter the nuclei of rat liver epithelial cells and cause caspase-independent apoptosis27. One intriguing potential biological role of DNase II–like proteins in trichocephalid parasites is the degradation of host DNA that may be released when the parasites damage host tissue during invasion, thereby limiting inflammatory and immune responses that could be elicited by undigested host DNA22.
(a) Normalized transcript levels of the 18 genes encoding DNase II domain–containing proteins in T. muris, comparing the parasite anterior region with the posterior regions of adult female and male parasites. Indication of significant transcriptional upregulation in a particular pairwise comparison (Up) refers to FDR % 0.01 and FDR > 1 × 10−5 when denoted by one asterisk and to FDR % 1 × 10−5 when denoted by two asterisks. For a full version of this figure, see Supplementary Figure 4b. (b) A maximum-likelihood phylogeny of DNase II protein domains (IPR004947) shows the relationships between the DNase II domains of proteins from Trichuris species, T. spiralis, other nematodes, insects and other invertebrates, and vertebrates. See Supplementary Figure 6 for a fully annotated version of this tree. The circled numbers highlight individual sequences of particular interest: (1) TMUE_s0085001500 1–358 (T. muris), TTRE_0000372701 1–317 (T. trichiura) and E5SXW8_TRISP 8–361 (UniProt E5SXW8); (2) TMUE_s0015002900 1–191 and TTRE_0000937801 31–255; and (3) E5S4S7_TRISP 14–306 (UniProt E5S4S7).
We found male-specific expression (Supplementary Table 8) of transcripts encoding proteins with major sperm protein (MSP) domains, which likely have a role in the amoeboid locomotion of nematode sperm28, and proteins with casein kinase–related and epidermal growth factor (EGF)-like domains, which might be involved in male mating-associated functions29. In contrast, transcripts for proteins with chitin-binding domains were upregulated in female whipworms. As nematode eggshells commonly contain chitin, it is likely that these proteins are associated with the formation of the eggshell or a related function30. The transcriptional landscape of L2 and L3 larvae is similar to that of the anterior region of adult worms, likely owing both to the shared intraepithelial location and the shared absence of the reproduction- and sex-specific transcripts that dominate the posterior region of adult worms (Supplementary Table 8). In addition, larval stages showed high expression of transcripts for ribosomal proteins and collagen- and fibronectin-related proteins, likely reflecting the fast growth and associated protein and cuticle synthesis in whipworms during the larval stage (Supplementary Fig. 4 and Supplementary Tables 8 and 9).
Host intestinal response to infection
Low-dose infection of mice with T. muris leads to chronic infection, whereas high-dose infections are typically cleared in the majority of inbred mouse strains. Therefore, low-dose infection of C57BL/6 mice with T. muris is used as a model of natural chronic infection of humans with T. trichiura, which usually presents with a low parasite burden31, and may also provide a model for inflammatory immune diseases of the gut32,33. A TH2 response is required for the resolution of infection and the acquisition of immunity34, but chronic infections are typified by a regulated TH1 response35,36. This combined response favors the parasite, counteracting a protective TH2 response, resulting in slower epithelial turnover and leading to pathology with features of colitis while preventing severe intestinal pathology37. To investigate the precise nature of these responses, we characterized gene expression changes in mouse cecum and mesenteric lymph node (MLN) upon infection, using RNA sequencing (RNA-seq). Changes in host gene expression were shared by the precise site of worm occupation and worm-free areas. We found only five genes differentially expressed in these tissues in infected mice compared to control mice, suggesting broad regulatory activity within the cecum.
Within infected cecum, we found 868 genes upregulated and 590 downregulated compared to the same tissue in naive mice. Upregulated genes were enriched for a variety of functional terms associated with the immune system, suggesting that these factors are the primary mediators of interaction with the worm during chronic infection. As expected, these changes were consistent with a TH1 response: Cd4 expression was upregulated by 4-fold, whereas proinflammatory cytokines Ifng (encoding interferon (IFN)-γ) and Tnfa (encoding tumor necrosis factor (TNF)-α) were upregulated by 26- and 12-fold, respectively (Supplementary Fig. 7). Conversely, Il4, Il5, Il9 and Il13, encoding cytokines typical of a TH2 response, were not differentially expressed. IL-18 has been suggested to have a role in promoting susceptibility by inhibiting the TH2 response38; however, at this stage of infection, we saw a downregulation of Il18 levels in susceptible mice, suggesting instead that this cytokine has a role in downregulating an excessive type 1 response. We observed upregulation of Il16 and Ccr5, which have been implicated in TH1 cell migration39 but have not previously been associated with Trichuris infection. There was strong upregulation of immunoglobulin classes G2B, G2C, A and M but not E in the cecum after infection, reflecting changes seen in MLN (Supplementary Fig. 7 and Supplementary Table 10). These changes in the upregulation of immunoglobulin classes likely arise as a result of IFN-γ stimulation, but a role for these isotypes in chronic infection is unknown. Certainly, antibody per se is thought to be dispensable for protection against primary infections40. In chronic infections, the antibody response seen here might be a non-functional byproduct of a parasite-induced TH1 response or might instead be a response to bacterial infection associated with parasite-induced intestinal damage. Support for the latter hypothesis is provided by the observed secretory IgA response, which is commonly associated with commensal bacteria41. The full extent of differential expression of cytokines and chemokines in infected cecum is shown in Supplementary Table 11. In MLN, 381 genes were upregulated and 176 genes were downregulated after infection. The gene set was dominated by genes related to the cell cycle and IgG, suggesting prolonged production of B cells. Whether these changes in expression simply indicate a response to chronic intestinal infection or increased exposure to microflora or suggest that these B cells or isotypes have specific or new functional roles remains to be determined.
In a chronic infection, it is essential that the parasite limits damage to the host. In particular, IL-10 has been proposed to control tissue repair during chronic infection35. Furthermore, IL-22, another member of the IL-10 superfamily, has been implicated in therapeutic T. trichiura infection42. We observed that Il10 and Il22 levels were upregulated in infected cecum. The source of these cytokines could be the classically induced FoxP3+ regulatory T (Treg) cells; however, we did not observe upregulation of mRNA for the associated markers FOXP3 and CD25, suggesting that the source might be other IL-10–producing T cell subsets. Mucosal mast cells have previously been observed in acute infection with T. muris but do not appear to be responsible for parasite resistance43. Evidence from the AKR mouse model of chronic infection44 and here from the low-dose C57BL/6 model suggests that these cells increase in number during chronic infection and therefore might have a role in tissue repair or immunoregulation, perhaps through the production of IL-10 (Supplementary Fig. 7). We observed expression of several markers of M2 (alternatively activated) macrophages (Supplementary Fig. 7), which have also been shown to be dispensable for resistance to Trichuris infection45. These macrophages are thought to be specific to a TH2 response, and, although we did not detect upregulation of Il4 or Il13, very low levels of these cytokines could be present, as we detected no reads for their transcripts in uninfected samples and low numbers of reads in infected samples. M2 macrophages are known to have roles in tissue repair46, and their dispensability for parasite clearance and presence during chronic infection indicate that this might be their function during Trichuris infection.
The pathogenesis caused by T. muris infection of C57BL/6 mice is similar to that seen in human ulcerative colitis. Furthermore, by crossing mice resistant and susceptible to T. muris infection, quantitative trait loci (QTLs) have been identified that are shared with other experimental models of colitis32. Here we showed that T. muris infection causes changes in the expression of genes associated through genome-wide association studies (GWAS) with ulcerative colitis and several other inflammatory diseases (Supplementary Fig. 8 and Supplementary Table 12). These findings provide support for the use of T. muris as a model of these devastating diseases prevalent in countries that are relatively free of intestinal parasites, in addition to its role as a model of T. trichiura infection.
Insights into new drug targets
Recent meta-analyses have showed that albendazole and mebendazole, used to treat trichuriasis, may only completely clear about one-third of infections, and high rates of reinfection are commonly encountered47,48. With the unsatisfactory performance of current anthelmintics and the availability of complete genome sequences, new opportunities for target-based approaches to drug discovery need to be explored. The preceding sections of this study highlight parasite processes that could potentially be disrupted by drugs (for example, protease, protease inhibitor and DNase II activities), but, by combining our transcriptome data with results from database searches, we have assigned, on a genome-wide scale, potentially desirable properties for drug target selection. These properties can be weighted and ranked (Supplementary Tables 13 and 14) or simply filtered. For instance, 8,307 genes are expressed in adult whipworms, 4,269 genes have high inferred druggability and, for 600 genes, we inferred evidence of essentiality. We found only 397 putative targets that fulfilled all these criteria. This list was further reduced to 29 genes by filtering for candidate proteins with homologs that are targets of existing approved drugs (Table 2 and Supplementary Tables 13 and 14). Among these, we identified fatty acid synthase, the target of the anti-obesity drug orlistat; DNA topoisomerase, a target for the previously used antischistosomiasis drug lucanthone; and calmodulin, the target for the antidiarrheal drug loperamide, which has previously been reported to increase the efficacy of mebendazole in treating trichuriasis49.
Discussion
Whipworms are exquisitely well adapted to their unusual biological niche. They are able to invade and maintain over extended periods of time their position within the epithelium of the large intestine, one of the fastest renewing tissues of the body, often without causing excessive pathology. The anterior end of adult worms appears to be key for host-parasite interactions, immunomodulation and feeding. This is because it is in intimate contact with host tissue and because it is home to two specialized cellular structures—the bacillary band and the stichosome. These adaptations are unique to whipworms and related parasites, that is, organisms that spend part of their life cycle buried in the intestinal mucosa of the host. This study identifies numerous genes with anterior end–specific expression that encode factors including protease inhibitors, DNase II and serine proteases. Many of these molecules are likely secreted from the worm and are plausibly involved in both immunomodulation and feeding. How and where secreted proteins are actually released from adult worms remains a fundamental and unanswered question; they could either be routed via the esophagus and exit via the anus into the intestinal lumen or find their way to the exterior of the worm via the intriguing pores of the bacillary cells that are in direct contact with host cell cytoplasm50. It is also unknown whether feeding actually occurs via the mouth, as Trichuris lacks the muscular pharynx of other nematodes that pump food through the gut. Alternatively, whipworms could both secrete digestive enzymes (together with immunomodulatory molecules) and absorb nutritional molecules via the pores of bacillary cells.
Here we also present the landscape of transcriptomic changes in chronically infected host tissue and suggest aspects of the host immune system that might be involved in reducing pathology. Whether this reduction in pathology is mediated by alternatively activated macrophages, mast cells or other cell populations, determining how the interplay of host and parasite regulates the immune system will also help in gaining a better understanding of diseases such as ulcerative colitis that appear to have much in common with whipworm infection. Intriguingly, the pig whipworm Trichuris suis is licensed as a medicine for treating inflammatory bowel diseases. Comparative immunology of these infections should help in developing more targeted therapies in the future. Finally, this work emphasizes the power of combining genomics and transcriptomics to study an important human pathogen and its well-defined model system in tandem, in not only opening up novel avenues of future therapeutics in human disease but also in providing fundamental biological data toward understanding of the host-parasite relationship.
Methods
Methods and materials are described here in brief. For further details, see the Supplementary Note. All animal experiments were performed under the auspices of the University of Manchester ethical review committee and under the UK Home Office Scientific Procedures Act (1986).
Genome sequencing.
Whipworm genome sequences are based on a single male individual of T. trichiura from Ecuador and a pool of T. muris parasites of the Edinburgh strain grown in male athymic nude mice that were 6–12 weeks of age and bred in the Biological Services Unit at the University of Manchester. For T. trichiura, the study protocol was approved by the ethics committees of the Hospital Pedro Vicente Maldonado, Pichincha Province, and Pontificia Universidad Católica del Ecuador, Quito, Ecuador, and included appropriate informed written consent. Genome sequencing was performed using both Illumina (T. trichiura and T. muris) and 454 (T. muris) platforms. To generate separate male and female T. muris samples (1 male individual and a pool of 11 females), worms were separated by sex on the basis of size and morphology. Sequenced libraries are listed in Supplementary Table 15.
Genome assembly and improvement.
Genome assembly was carried out with SGA52 v0.9.17 and Velvet53 v1.2.03 (T. trichiura) as well as Celera54 v7.0 (T. muris). Misassemblies were identified and corrected using REAPR55 and manual inspection. Genome assemblies were scaffolded and improved using SSPACE56, IMAGE57, Gapfiller58 and (for T. muris) by incorporating optical map data with tools from OpGen. For T. muris, genome assembly v2.1 was used to predict genes, whereas assembly v4 represents the most recent assembly with the best long-range contiguation for which sequence scaffolds were joined also in cases where the corresponding gaps were too large to be spanned by sequencing reads (larger than ∼10 kb) but were confidently spanned by optical map contigs.
Transcriptome sequencing—T. muris and mouse.
High-throughput transcriptome data were generated from the RNA of T. muris and mouse tissue (14 male C57BL/6 mice infected at 6–8 weeks of age). For larval stage and adult whipworms, RNA was prepared using TRIzol and lysing matrix D (1.4-mm ceramic spheres) and a Fastprep24 (MP Biomedicals). Mice had been subjected to a low-dose infection with T. muris (25 eggs by oral gavage) or were uninfected, and the sampled tissues included mesenteric lymph node, a section of cecum where the worms reside and—as a control—an uninfected section of cecum. RNA-seq libraries were prepared following the RNA-seq protocols of the Illumina mRNA-Seq Sample Prep kit and the Illumina TruSeq kit. Transcriptome libraries were sequenced on Illumina HiSeq 2000 machines and are listed in Supplementary Table 16.
Gene predictions.
For T. muris, the gene predictor Augustus59 v2.4 was trained on a gene training set—derived from CEGMA6 predictions, Augustus predictions and manual curation—and was run by also providing intron hints on the basis of RNA-seq data. Genes of particular interest and genes that appeared incorrect on the basis of semiautomatic screens and manual inspection were manually curated. Likely transposon-related genes were removed from the final gene set. For T. trichiura, genes were predicted using a Maker60 v2.2.28 pipeline that incorporated ab initio predictions by Augustus, GeneMark-ES61 v2.3a (self-trained) and SNAP62 2013-02-16 and considered gene models on the basis of comparison with other species.
Functional gene annotation.
Functional gene annotation including the assignment of gene product descriptions and GO terms was based on Interpro protein domain analysis and BLAST searches against annotated genomes. Pfam protein domain predictions and GO term assignments as provided by InterproScan are listed in Supplementary Tables 17 and 18. Genes lacking both Pfam domain and BLAST hit were labeled 'hypothetical protein'. For T. trichiura, genes with a one-to-one ortholog in T. muris, functional annotation was transferred from the T. muris ortholog.
Gene family clustering and phylogenetic analysis.
Gene families were determined using orthoMCL63 v2.0, and one-to-one orthologs were identified with Inparanoid64 v4.1 and OMA65 v0.99t. Multiple-sequence alignments were created using MAFFT66 v6.857 and GBlocks67 v0.91b, and phylogenetic trees were constructed with RAxMLHPC68 v7.2.8. Sequences to be included in the phylogenetic tree for DNase II were selected on the basis of the presence of the corresponding Interpro protein domain IPR004947.
Chromosome-level analysis.
The assignment of scaffolds and genes to chromosomes was performed by first clustering the numbers of one-to-one orthologs in the largest genome assembly scaffolds of T. muris and T. spiralis, which yielded three distinct 'linkage groups'. More scaffolds of T. muris could then be assigned to linkage groups on the basis of the presence of gene orthologs in comparison to the large T. spiralis scaffolds that were already assigned to a linkage group. Sequence read coverage was determined by aligning Illumina data against the relevant genome assembly using SMALT v0.7.4 followed by running the command genomecov of BEDTools69 v2.17.0. Heterozygosity per 10-kb window of genomic sequence was calculated on the basis of a pileup including base and variant calls that was generated by SAMtools70 mpileup v0.1.18 from the high-throughput sequence read alignment. Genomic scaffolds representing the X chromosome were identified on the basis of both read coverage and heterozygosity. Mean read coverage over parts of the genome assembly with extraordinarily high read coverage was also used to estimate the real extent of DNA content in the parasite represented by such parts of the assembly. Centromeric sequences were putatively identified on the basis of high read coverage and the length of the underlying repeat units (164 bp to 176 bp).
Gene expression analysis—T. muris and mouse.
For differential gene expression analysis, paired-end Illumina RNA-seq data were mapped using TopHat71 v1.4.1 to either the T. muris genome v2.1 (for T. muris samples) or a combined reference of mouse (mm10) and T. muris transcripts (for mouse samples). The number of reads per gene were determined with BEDTools (T. muris) or eXpress72 (mouse), and analysis of differential expression was carried out using the Bioconductor packages edgeR73 v3.2.4 (T. muris) and DESeq74 (mouse). GO term enrichment analysis was carried out with TopGO75 and innateDB76, and protein domain enrichment analysis was based on the results of Interproscan77.
Identification of new drug targets.
The ranking of all T. muris proteins for their suitability as a drug target was based on a number of different types of information: T. muris RNA-seq expression data; orthology (as determined by OMA) of T. muris protein sequences to those in T. trichiura, C. elegans, mouse, human and drug targets; protein homolog essentiality in mouse and C. elegans; whether a protein was predicted to be an enzyme, on the basis of KEGG orthology; druggability information from ChEMBL; and drug target information from DrugBank and from TTD (Therapeutic Targets Database). Nutraceutical targets were filtered out.
GWAS analysis.
A test to identify genes that were both differentially expressed in the cecum of whipworm-infected versus uninfected mice and that were associated with immune-mediated diseases was performed as follows. Mouse genes were filtered for those that had a unique human ortholog, were annotated as protein coding and were located on autosomes. Lists of associated loci from published GWAS were extracted for four immune-mediated complex diseases (Crohn's disease, ulcerative colitis, celiac disease and type 1 diabetes), as well as for two likely immune-unrelated complex traits (height and body mass index). Testing for enrichment was performed using a Monte Carlo simulation approach that accounts for linkage disequilibrium between associated SNPs and non-random arrangement of functionally related genes within the genome.
URLs.
MEROPS peptidase database, http://merops.sanger.ac.uk/.
Accession codes.
The Trichuris genome projects have been registered under the International Nucleotide Sequence Database Collaboration (INSDC) project ID numbers PRJEB2108 (T. muris) and PRJEB2679 (T. trichiura). Genome data and the complete genome annotation are available for download from ftp://ftp.sanger.ac.uk/pub/pathogens/Trichuris/GenomePaper2014. Genome sequencing data have been deposited in the European Nucleotide Archive (ENA) under sample accession numbers ERS016744, ERS016965, ERS056020 and ERS244696 for T. muris and under sample accession number ERS056020 for T. trichiura. RNA-seq transcriptome data have been deposited in ENA under study accession numbers ERP002000 (T. muris) and ERP002560 (mouse) and in ArrayExpress under study accession numbers E-ERAD-125 (T. muris) and E-ERAD-181 (mouse).
Accession codes
Primary accessions
ArrayExpress
BioProject
European Nucleotide Archive
References
Bethony, J. et al. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367, 1521–1532 (2006).
Grencis, R.K. & Bancroft, A.J. Interleukin-13: a key mediator in resistance to gastrointestinal-dwelling nematode parasites. Clin. Rev. Allergy Immunol. 26, 51–60 (2004).
Summers, R.W. et al. Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. Am. J. Gastroenterol. 98, 2034–2041 (2003).
Jouvin, M.H. & Kinet, J.P. Trichuris suis ova: testing a helminth-based therapy as an extension of the hygiene hypothesis. J. Allergy Clin. Immunol. 130, 3–10, quiz 11–12 (2012).
Parkinson, J. et al. A transcriptomic analysis of the phylum Nematoda. Nat. Genet. 36, 1259–1267 (2004).
Parra, G., Bradnam, K. & Korf, I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007).
Spakulová, M., Kralova, I. & Cutillas, C. Studies on the karyotype and gametogenesis in Trichuris muris. J. Helminthol. 68, 67–72 (1994).
Mutafova, T., Dimitrova, Y. & Komandarev, S. The karyotype of four Trichinella species. Z. Parasitenkd. 67, 115–120 (1982).
Desjardins, C.A. et al. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat. Genet. 45, 495–500 (2013).
Kikuchi, T. et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7, e1002219 (2011).
Mitreva, M. et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 43, 228–235 (2011).
Ghedin, E. et al. Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 (2007).
Hasnain, S.Z., McGuckin, M.A., Grencis, R.K. & Thornton, D.J. Serine protease(s) secreted by the nematode Trichuris muris degrade the mucus barrier. PLoS Negl. Trop. Dis. 6, e1856 (2012).
Drake, L.J., Bianco, A.E., Bundy, D.A. & Ashall, F. Characterization of peptidases of adult Trichuris muris. Parasitology 109, 623–630 (1994).
Marcilla, A. et al. The transcriptome analysis of Strongyloides stercoralis L3i larvae reveals targets for intervention in a neglected disease. PLoS Negl. Trop. Dis. 6, e1513 (2012).
Cantacessi, C. et al. Massively parallel sequencing and analysis of the Necator americanus transcriptome. PLoS Negl. Trop. Dis. 4, e684 (2010).
Cantacessi, C. et al. The transcriptome of Trichuris suis—first molecular insights into a parasite with curative properties for key immune diseases of humans. PLoS ONE 6, e23590 (2011).
Bénard, C.Y., Boyanov, A., Hall, D.H. & Hobert, O. DIG-1, a novel giant protein, non-autonomously mediates maintenance of nervous system architecture. Development 133, 3329–3340 (2006).
Williams, S.E., Brown, T.I., Roghanian, A. & Sallenave, J.M. SLPI and elafin: one glove, many fingers. Clin. Sci. (Lond.) 110, 21–35 (2006).
Scott, A., Weldon, S. & Taggart, C.C. SLPI and elafin: multifunctional antiproteases of the WFDC family. Biochem. Soc. Trans. 39, 1437–1440 (2011).
Wilkinson, T.S., Roghanian, A., Simpson, A.J. & Sallenave, J.M. WAP domain proteins as modulators of mucosal immunity. Biochem. Soc. Trans. 39, 1409–1415 (2011).
Liu, M.F. et al. The functions of deoxyribonuclease II in immunity and development. DNA Cell Biol. 27, 223–228 (2008).
Mitreva, M. & Jasmer, D.P. Advances in the sequencing of the genome of the adenophorean nematode Trichinella spiralis. Parasitology 135, 869–880 (2008).
Bien, J. et al. Comparative analysis of excretory-secretory antigens of Trichinella spiralis and Trichinella britovi muscle larvae by two-dimensional difference gel electrophoresis and immunoblotting. Proteome Sci. 10, 10 (2012).
Crow, Y.J. The story of DNase II: a stifled death-wish leads to self-harm. Eur. J. Immunol. 40, 2376–2378 (2010).
Lai, H.J., Lo, S.J., Kage-Nakadai, E., Mitani, S. & Xue, D. The roles and acting mechanism of Caenorhabditis elegans DNase II genes in apoptotic DNA degradation and development. PLoS ONE 4, e7348 (2009).
Ota, E. et al. Caspase-independent apoptosis induced in rat liver cells by plancitoxin I, the major lethal factor from the crown-of-thorns starfish Acanthaster planci venom. Toxicon 48, 1002–1010 (2006).
Tarr, D.E. & Scott, A.L. MSP domain proteins. Trends Parasitol. 21, 224–231 (2005).
Hu, J., Bae, Y.K., Knobel, K.M. & Barr, M.M. Casein kinase II and calcineurin modulate TRPP function and ciliary localization. Mol. Biol. Cell 17, 2200–2211 (2006).
Johnston, W.L., Krizus, A. & Dennis, J.W. Eggshell chitin and chitin-interacting proteins prevent polyspermy in C. elegans. Curr. Biol. 20, 1932–1937 (2010).
Bancroft, A.J., Else, K.J. & Grencis, R.K. Low-level infection with Trichuris muris significantly affects the polarization of the CD4 response. Eur. J. Immunol. 24, 3113–3118 (1994).
Levison, S.E. et al. Genetic analysis of the Trichuris muris–induced model of colitis reveals QTL overlap and a novel gene cluster for establishing colonic inflammation. BMC Genomics 14, 127 (2013).
Levison, S.E. et al. Colonic transcriptional profiling in resistance and susceptibility to trichuriasis: phenotyping a chronic colitis and lessons for iatrogenic helminthosis. Inflamm. Bowel Dis. 16, 2065–2079 (2010).
Else, K.J., Finkelman, F.D., Maliszewski, C.R. & Grencis, R.K. Cytokine-mediated regulation of chronic intestinal helminth infection. J. Exp. Med. 179, 347–351 (1994).
Schopf, L.R., Hoffmann, K.F., Cheever, A.W., Urban, J.F. Jr. & Wynn, T.A. IL-10 is critical for host resistance and survival during gastrointestinal helminth infection. J. Immunol. 168, 2383–2392 (2002).
D'Elia, R., Behnke, J.M., Bradley, J.E. & Else, K.J. Regulatory T cells: a role in the control of helminth-driven intestinal pathology and worm survival. J. Immunol. 182, 2340–2348 (2009).
Cliffe, L.J. et al. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science 308, 1463–1465 (2005).
Helmby, H., Takeda, K., Akira, S. & Grencis, R.K. Interleukin (IL)-18 promotes the development of chronic gastrointestinal helminth infection by downregulating IL-13. J. Exp. Med. 194, 355–364 (2001).
Lynch, E.A., Heijens, C.A., Horst, N.F., Center, D.M. & Cruikshank, W.W. Cutting edge: IL-16/CD4 preferentially induces Th1 cell migration: requirement of CCR5. J. Immunol. 171, 4965–4968 (2003).
Else, K.J. & Grencis, R.K. Antibody-independent effector mechanisms in resistance to the intestinal nematode parasite Trichuris muris. Infect. Immun. 64, 2950–2954 (1996).
Bos, N.A., Jiang, H.Q. & Cebra, J.J. T cell control of the gut IgA response against commensal bacteria. Gut 48, 762–764 (2001).
Broadhurst, M.J. et al. IL-22+ CD4+ T cells are associated with therapeutic Trichuris trichiura infection in an ulcerative colitis patient. Sci. Transl. Med. 2, 60ra88 (2010).
Koyama, K. & Ito, Y. Mucosal mast cell responses are not required for protection against infection with the murine nematode parasite Trichuris muris. Parasite Immunol. 22, 13–20 (2000).
Datta, R. et al. Identification of novel genes in intestinal tissue that are regulated after infection with an intestinal nematode parasite. Infect. Immun. 73, 4025–4033 (2005).
Bowcutt, R. et al. Arginase-1–expressing macrophages are dispensable for resistance to infection with the gastrointestinal helminth Trichuris muris. Parasite Immunol. 33, 411–420 (2011).
Varin, A. & Gordon, S. Alternative activation of macrophages: immune function and cellular biology. Immunobiology 214, 630–641 (2009).
Jia, T.W., Melville, S., Utzinger, J., King, C.H. & Zhou, X.N. Soil-transmitted helminth reinfection after drug treatment: a systematic review and meta-analysis. PLoS Negl. Trop. Dis. 6, e1621 (2012).
Keiser, J. & Utzinger, J. Efficacy of current drugs against soil-transmitted helminth infections: systematic review and meta-analysis. J. Am. Med. Assoc. 299, 1937–1948 (2008).
Scragg, J.N. & Proctor, E.M. Further experience with mebendazole in the treatment of symptomatic trichuriasis in children. Am. J. Trop. Med. Hyg. 27, 255–257 (1978).
Tilney, L.G., Connelly, P.S., Guild, G.M., Vranich, K.A. & Artis, D. Adaptation of a nematode parasite to living within the mammalian epithelium. J. Exp. Zool. A Comp. Exp. Biol. 303, 927–945 (2005).
Knox, C. et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 39, D1035–D1041 (2011).
Simpson, J.T. & Durbin, R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 22, 549–556 (2012).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Myers, E.W. et al. A whole-genome assembly of Drosophila. Science 287, 2196–2204 (2000).
Hunt, M. et al. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14, R47 (2013).
Boetzer, M., Henkel, C.V., Jansen, H.J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011).
Tsai, I.J., Otto, T.D. & Berriman, M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 11, R41 (2010).
Nadalin, F., Vezzi, F. & Policriti, A. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics 13 (suppl. 14), S8 (2012).
Stanke, M., Schoffmann, O., Morgenstern, B. & Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62 (2006).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011).
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y.O. & Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18, 1979–1990 (2008).
Korf, I. Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004).
Li, L., Stoeckert, C.J. & Roos, D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
O'Brien, K.P., Remm, M. & Sonnhammer, E.L. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 33, D476–D480 (2005).
Altenhoff, A.M., Schneider, A., Gonnet, G.H. & Dessimoz, C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 39, D289–D294 (2011).
Katoh, K. & Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298 (2008).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Roberts, A. & Pachter, L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10, 71–73 (2013).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Alexa, A., Rahnenfuhrer, J. & Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006).
Lynn, D.J. et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 4, 218 (2008).
Quevillon, E. et al. InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120 (2005).
Acknowledgements
We thank R. Maizels for initially proposing a T. muris genome project, C. Anderson and T. Raine for sharing their unpublished method for enrichment analysis of IBD-associated genes, S. Thompson for preparing parasite material, T. Kikuchi and Z. Hance for preparing genomic DNA, H.M. Bennett for preparing RNA, K.L. Brooks and H. Beasley for manually curating gene models, M. Dunn for generating optical map data, N. Rawlings for helpful discussions, A. Coghlan for supporting bioinformatics analysis and M. Aslett for data submission to the European Molecular Biology Laboratory (EMBL). This work was funded by the Wellcome Trust through their support of the Wellcome Trust Sanger Institute (grant 098051) and through an Investigator Award (WT100290MA) and Programme grant (WT083620MA) to R.K.G. The collection of samples in Ecuador was supported by the Wellcome Trust (grant 088862/Z/09/Z).
Author information
Authors and Affiliations
Contributions
A.J.B. cultivated T. muris parasites in mice and prepared DNA and RNA. P.J.C. provided T. trichiura parasite material. T.H. prepared RNA samples. I.J.T. assembled the genomes and produced chromosomal assignments. A.T. and S.N. manually improved the genome assemblies and curated genes. B.J.F., E.J.S. and I.J.T. trained gene-finding software and structurally annotated the genomes. B.J.F. and M.Z. provided functional gene annotation and analysis. J.A.C. performed gene family clustering and analysis. B.J.F. analyzed the T. muris transcriptome, genome read coverage and heterozygosity. A.J.R. analyzed the mouse transcriptome. M.B. and M.Z. analyzed drug targets. N.H. coordinated the project and managed the sequencing. J.Z.L. carried out the GWAS analysis. A.J.B., A.J.R., B.J.F., M.B. and R.K.G. wrote the manuscript. M.B. and R.K.G. directed the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 2, 3, 11, 15 and 16, and Supplementary Note (PDF 8230 kb)
Supplementary Table 1
Chromosomal location. (XLSX 431 kb)
Supplementary Table 4
Protease and protease inhibitor content of Trichuris genomes (classification according to the MEROPS peptidase database). (XLSX 231 kb)
Supplementary Table 5
T. muris RNA-seq read counts and transcript abundance. (XLSX 3310 kb)
Supplementary Table 6
T. muris RNA-seq differential expression analysis. (XLSX 7237 kb)
Supplementary Table 7
T. muris gene groupings of interest. (XLSX 80 kb)
Supplementary Table 8
Protein domain enrichment analysis. (XLSX 62 kb)
Supplementary Table 9
GO term enrichment analysis. (XLSX 37 kb)
Supplementary Table 10
Mouse differential expression analysis. (XLSX 7410 kb)
Supplementary Table 12
Gene overlap. (XLSX 51 kb)
Supplementary Table 13
Highly ranked putative drug targets in Trichuris. (XLSX 32 kb)
Supplementary Table 14
Prediction of drug targets in Trichuris. (XLSX 3274 kb)
Supplementary Table 17
Pfam protein domain predictions for T. muris gene set v2.3 and T. trichiura gene set v2.2. (XLSX 2924 kb)
Supplementary Table 18
GO term assignment for T. muris gene set v2.3 and T. trichiura gene set v2.2. (XLSX 243 kb)
Supplementary Data Set 1
OrthoMCL clustering. (TXT 3837 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/.
About this article
Cite this article
Foth, B., Tsai, I., Reid, A. et al. Whipworm genome and dual-species transcriptome analyses provide molecular insights into an intimate host-parasite interaction. Nat Genet 46, 693–700 (2014). https://doi.org/10.1038/ng.3010
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3010
This article is cited by
-
A genome-scale metabolic model of parasitic whipworm
Nature Communications (2023)
-
Whipworm secretions and their roles in host-parasite interactions
Parasites & Vectors (2022)
-
Population genomics of ancient and modern Trichuris trichiura
Nature Communications (2022)
-
Evolution of sexual systems, sex chromosomes and sex-linked gene transcription in flatworms and roundworms
Nature Communications (2022)
-
Whip-LAMP: a novel LAMP assay for the detection of Trichuris muris-derived DNA in stool and urine samples in a murine experimental infection model
Parasites & Vectors (2020)