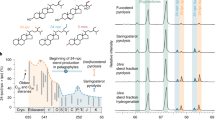
Abstract
Natural products preserved in the geological record can function as ‘molecular fossils’, providing insight into organisms and physiologies that existed in the deep past. One important group of molecular fossils is the steroidal hydrocarbons (steranes), which are the diagenetic remains of sterol lipids. Complex sterols with modified side chains are unique to eukaryotes, although simpler sterols can also be synthesized by a few bacteria1. Sterol biosynthesis is an oxygen-intensive process; thus, the presence of complex steranes in ancient rocks not only signals the presence of eukaryotes, but also aerobic metabolic processes2. In 1999, steranes were reported in 2.7 billion year (Gyr)-old rocks from the Pilbara Craton in Australia3, suggesting a long delay between photosynthetic oxygen production and its accumulation in the atmosphere (also known as the Great Oxidation Event) 2.45–2.32 Gyr ago4. However, the recent reappraisal and rejection of these steranes as contaminants5 pushes the oldest reported steranes forward to around 1.64 Gyr ago (ref. 6). Here we use a molecular clock approach to improve constraints on the evolution of sterol biosynthesis. We infer that stem eukaryotes shared functionally modern sterol biosynthesis genes with bacteria via horizontal gene transfer. Comparing multiple molecular clock analyses, we find that the maximum marginal probability for the divergence time of bacterial and eukaryal sterol biosynthesis genes is around 2.31 Gyr ago, concurrent with the most recent geochemical evidence for the Great Oxidation Event7. Our results therefore indicate that simple sterol biosynthesis existed well before the diversification of living eukaryotes, substantially predating the oldest detected sterane biomarkers (approximately 1.64 Gyr ago6), and furthermore, that the evolutionary history of sterol biosynthesis is tied to the first widespread availability of molecular oxygen in the ocean–atmosphere system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wei, J. H., Yin, X. & Welander, P. V. Sterol synthesis in diverse bacteria. Front. Microbiol. 7, 990 (2016)
Summons, R. E., Bradley, A. S., Jahnke, L. L. & Waldbauer, J. R. Steroids, triterpenoids and molecular oxygen. Philos. Trans. R. Soc. B Biol. Sci. 361, 951–968 (2006)
Brocks, J. J., Logan, G. A., Buick, R. & Summons, R. E. Archean molecular fossils and the early rise of eukaryotes. Science 285, 1033–1036 (1999)
Bekker, A. et al. Dating the rise of atmospheric oxygen. Nature 427, 117–120 (2004)
French, K. L. et al. Reappraisal of hydrocarbon biomarkers in Archean rocks. Proc. Natl Acad. Sci. USA 112, 5915–5920 (2015)
Brocks, J. J. et al. Biomarker evidence for green and purple sulphur bacteria in a stratified Palaeoproterozoic sea. Nature 437, 866–870 (2005)
Luo, G. et al. Rapid oxygenation of Earth’s atmosphere 2.33 billion years ago. Sci. Adv. 2, e1600134 (2016)
Desmond, E. & Gribaldo, S. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 1, 364–381 (2009)
He, D. et al. An alternative root for the eukaryote tree of life. Curr. Biol. 24, 465–470 (2014)
Derelle, R. et al. Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl Acad. Sci. USA 112, E693–E699 (2015)
Pearson, A., Budin, M. & Brocks, J. J. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus . Proc. Natl Acad. Sci. USA 100, 15352–15357 (2003)
Douzery, E. J. P., Snell, E. A., Bapteste, E., Delsuc, F. & Philippe, H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl Acad. Sci. USA 101, 15386–15391 (2004)
Berney, C. & Pawlowski, J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. Biol. Sci. 273, 1867–1872 (2006)
Parfrey, L. W., Lahr, D. J. G., Knoll, A. H. & Katz, L. A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl Acad. Sci. USA 108, 13624–13629 (2011)
Farquhar, J., Bao, H. & Thiemens, M. Atmospheric influence of Earth’s earliest sulfur cycle. Science 289, 756–759 (2000)
Lyons, T. W., Reinhard, C. T. & Planavsky, N. J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014)
Cavalier-Smith, T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297–354 (2002)
Illing, C. J., Hallmann, C., Miller, K. E., Summons, R. E. & Strauss, H. Airborne hydrocarbon contamination from laboratory atmospheres. Org. Geochem. 76, 26–38 (2014)
Summons, R. E. et al. Distinctive hydrocarbon biomarkers from fossiliferous sediment of the Late Proterozoic Walcott Member, Chuar Group, Grand Canyon, Arizona. Geochim. Cosmochim. Acta 52, 2625–2637 (1988)
Love, G. D., Stalvies, C., Grosjean, E., Meredith, W. & Snape, C. E. in Paleontological Society Papers Vol. 14 (eds Kelley, P. H. & Bambach, R. K. ) 67–83 (The Paleontological Society, 2008)
Brocks, J. J. et al. Early sponges and toxic protists: possible sources of cryostane, an age diagnostic biomarker antedating Sturtian Snowball Earth. Geobiology 14, 129–149 (2016)
Gold, D. A. et al. Sterol and genomic analyses validate the sponge biomarker hypothesis. Proc. Natl Acad. Sci. USA 113, 2684–2689 (2016)
Banta, A. B., Wei, J. H. & Welander, P. V. A distinct pathway for tetrahymanol synthesis in bacteria. Proc. Natl Acad. Sci. USA 112, 13478–13483 (2015)
Anbar, A. D. & Knoll, A. H. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297, 1137–1142 (2002)
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016)
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006)
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003)
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012)
Yin, Z. et al. Sponge grade body fossil with cellular resolution dating 60 Myr before the Cambrian. Proc. Natl Acad. Sci. USA 112, E1453–E1460 (2015)
Benton, M. J. et al. Constraints on the timescale of animal evolutionary history. Palaeontologia Electronica 18, 1–116 (2015)
Gold, D. A., Runnegar, B., Gehling, J. G. & Jacobs, D. K. Ancestral state reconstruction of ontogeny supports a bilaterian affinity for Dickinsonia. Evol. Dev. 17, 315–324 (2015)
Battistuzzi, F. U., Billing-Ross, P., Murillo, O., Filipski, A. & Kumar, S. A Protocol for diagnosing the effect of calibration priors on posterior time estimates: a case study for the Cambrian explosion of animal phyla. Mol. Biol. Evol. 32, 1907–1912 (2015)
Acknowledgements
We gratefully acknowledge funding from the Agouron Institute Geobiology Fellowship to D.A.G. and the Simons Foundation Collaboration on the Origins of Life to R.E.S. and G.P.F. Additional support was provided by the National Science Foundation programme ‘Frontiers of Earth System Dynamics’ (EAR-1338810) to R.E.S., and the National Science Foundation programme ‘Integrated Earth Systems’ (IES-1615426) to G.P.F.
Author information
Authors and Affiliations
Contributions
R.E.S. and D.A.G. designed the experiment. D.A.G. and A.C. performed the data analysis. All authors were involved in interpreting the data and drafting the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Supplementary information
Supplementary Information
This file contains Supplementary Results and Discussion and additional references. (PDF 346 kb)
Supplementary Data
This zipped file contains the files for Supplementary Data 1 and 2. In Data 1 all amino acid alignments and trees from this study are shown; the GenInfo Identifier (GI) numbers for sequences used are included in the taxon IDs. Data 2 contains the code used in this analysis. Please note that the authors place no restriction on its use. (ZIP 485 kb)
Rights and permissions
About this article
Cite this article
Gold, D., Caron, A., Fournier, G. et al. Paleoproterozoic sterol biosynthesis and the rise of oxygen. Nature 543, 420–423 (2017). https://doi.org/10.1038/nature21412
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21412
This article is cited by
-
Reconciling discrepant minor sulfur isotope records of the Great Oxidation Event
Nature Communications (2023)
-
Lost world of complex life and the late rise of the eukaryotic crown
Nature (2023)
-
Infancy of sterol biosynthesis hints at extinct eukaryotic species
Nature (2023)
-
De novo cholesterol biosynthesis in bacteria
Nature Communications (2023)
-
What are the Limiting Environmental Factors for the Evolution of Early Eukaryotic Diversity?
Journal of Earth Science (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.