Abstract
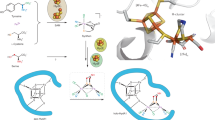
Membrane-bound respiratory [NiFe]-hydrogenase (MBH), a H2-uptake enzyme found in the periplasmic space of bacteria, catalyses the oxidation of dihydrogen: H2 → 2H+ + 2e− (ref. 1). In contrast to the well-studied O2-sensitive [NiFe]-hydrogenases (referred to as the standard enzymes), MBH has an O2-tolerant H2 oxidation activity2,3,4; however, the mechanism of O2 tolerance is unclear5. Here we report the crystal structures of Hydrogenovibrio marinus MBH in three different redox conditions at resolutions between 1.18 and 1.32 Å. We find that the proximal iron-sulphur (Fe-S) cluster of MBH has a [4Fe-3S] structure coordinated by six cysteine residues—in contrast to the [4Fe-4S] cubane structure coordinated by four cysteine residues found in the proximal Fe-S cluster of the standard enzymes—and that an amide nitrogen of the polypeptide backbone is deprotonated and additionally coordinates the cluster when chemically oxidized, thus stabilizing the superoxidized state of the cluster. The structure of MBH is very similar to that of the O2-sensitive standard enzymes except for the proximal Fe-S cluster. Our results give a reasonable explanation why the O2 tolerance of MBH is attributable to the unique proximal Fe-S cluster; we propose that the cluster is not only a component of the electron transfer for the catalytic cycle, but that it also donates two electrons and one proton crucial for the appropriate reduction of O2 in preventing the formation of an unready, inactive state of the enzyme.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vignais, P. M. & Billoud, B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272 (2007)
Vincent, K. A. et al. Electrocatalytic hydrogen oxidation by an enzyme at high carbon monoxide or oxygen levels. Proc. Natl Acad. Sci. USA 102, 16951–16954 (2005)
Luo, X., Brugna, M., Tron-Infossi, P., Giudici-Orticoni, M. T. & Lojou, E. Immobilization of the hyperthermophilic hydrogenase from Aquifex aeolicus bacterium onto gold and carbon nanotube electrodes for efficient H2 oxidation. J. Biol. Inorg. Chem. 14, 1275–1288 (2009)
Lukey, M. J. et al. How Escherichia coli is equipped to oxidize hydrogen under different redox conditions. J. Biol. Chem. 285, 3928–3938 (2010)
De Lacey, A. L., Fernandez, V. M., Rousset, M. & Cammack, R. Activation and inactivation of hydrogenase function and the catalytic cycle: spectroelectrochemical studies. Chem. Rev. 107, 4304–4330 (2007)
Brugna-Guiral, M. et al. [NiFe] hydrogenases from the hyperthermophilic bacterium Aquifex aeolicus: properties, function, and phylogenetics. Extremophiles 7, 145–157 (2003)
Burgdorf, T. et al. [NiFe]-hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygen-tolerant biological hydrogen oxidation. J. Mol. Microbiol. Biotechnol. 10, 181–196 (2005)
Yoon, K.-S., Fukuda, K., Fujisawa, K. & Nishihara, H. Purification and characterization of a highly thermostable, oxygen-resistant, respiratory [NiFe]-hydrogenase from a marine, aerobic hydrogen-oxidizing bacterium Hydrogenovibrio marinus. Int. J. Hydrogen Energy 36, 7081–7088 (2011)
Volbeda, A. et al. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373, 580–587 (1995)
Higuchi, Y., Yagi, T. & Yasuoka, N. Unusual ligand structure in Ni-Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis. Structure 5, 1671–1680 (1997)
Montet, Y. et al. Gas access to the active site of Ni-Fe hydrogenases probed by X-ray crystallography and molecular dynamics. Nature Struct. Biol. 4, 523–526 (1997)
Matias, P. M. et al. [NiFe] hydrogenase from Desulfovibrio desulfuricans ATCC 27774: gene sequencing, three-dimensional structure determination and refinement at 1.8 Å and modelling studies of its interaction with the tetrahaem cytochrome c3 . J. Biol. Inorg. Chem. 6, 63–81 (2001)
Fontecilla-Camps, J. C., Volbeda, A., Cavazza, C. & Nicolet, Y. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem. Rev. 107, 4273–4303 (2007)
Saggu, M. et al. Spectroscopic insights into the oxygen-tolerant membrane-associated [NiFe] hydrogenase of Ralstonia eutropha H16. J. Biol. Chem. 284, 16264–16276 (2009)
Peters, J. W. et al. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36, 1181–1187 (1997)
Berrisford, J. M. & Sazanov, L. A. Structural basis for the mechanism of respiratory complex I. J. Biol. Chem. 284, 29773–29783 (2009)
Aragão, D., Mitchell, E. P., Frazao, C. F., Carrondo, M. A. & Lindley, P. F. Structural and functional relationships in the hybrid cluster protein family: structure of the anaerobically purified hybrid cluster protein from Desulfovibrio vulgaris at 1.35 Å resolution. Acta Crystallogr. D 64, 665–674 (2008)
Huang, W. et al. Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold. Structure 5, 691–699 (1997)
Pandelia, M. E. et al. Characterization of a unique [FeS] cluster in the electron transfer chain of the oxygen tolerant [NiFe] hydrogenase from Aquifex aeolicus. Proc. Natl Acad. Sci. USA 108, 6097–6102 (2011)
Schneider, K., Patil, D. S. & Cammack, R. ESR properties of membrane-bound hydrogenase from aerobic hydrogen bacteria. Biochim. Biophys. Acta 748, 353–361 (1983)
Knüttel, K. et al. Redox properties of the metal centres in the membrane-bound hydrogenase from Alcaligenes eutrophus CH34. Bull. Pol. Acad. Sci. Chem. 42, 495–511 (1994)
Goris, T. et al. A unique iron-sulfur cluster is crucial for oxygen tolerance of a [NiFe]-hydrogenase. Nature Chem. Biol. 7, 310–318 (2011)
Dementin, S. et al. A glutamate is the essential proton transfer gate during the catalytic cycle of the [NiFe] hydrogenase. J. Biol. Chem. 279, 10508–10513 (2004)
Teixeira, V. H., Soares, C. M. & Baptista, A. M. Proton pathways in a [NiFe]-hydrogenase: A theoretical study. Proteins 70, 1010–1022 (2008)
Lubitz, W., Reijerse, E. & van Gastel, M. [NiFe] and [FeFe] hydrogenases studied by advanced magnetic resonance techniques. Chem. Rev. 107, 4331–4365 (2007)
Fernandez, V. M., Hatchikian, E. C., Patil, D. S. & Cammack, R. ESR-detectable nickel and iron-sulphur centres in relation to the reversible activation of Desulfovibrio gigas hydrogenase. Biochim. Biophys. Acta 883, 145–154 (1986)
Cracknell, J. A., Wait, A. F., Lenz, O., Friedrich, B. & Armstrong, F. A. A kinetic and thermodynamic understanding of O2 tolerance in [NiFe]-hydrogenases. Proc. Natl Acad. Sci. USA 106, 20681–20686 (2009)
Marques, M. C., Coelho, R., De Lacey, A. L., Pereira, I. A. & Matias, P. M. The three-dimensional structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough: a hydrogenase without a bridging ligand in the active site in its oxidised, “as-isolated” state. J. Mol. Biol. 396, 893–907 (2010)
Buhrke, T., Lenz, O., Krauss, N. & Friedrich, B. Oxygen tolerance of the H2-sensing [NiFe] hydrogenase from Ralstonia eutropha H16 is based on limited access of oxygen to the active site. J. Biol. Chem. 280, 23791–23796 (2005)
Duché, O., Elsen, S., Cournac, L. & Colbeau, A. Enlarging the gas access channel to the active site renders the regulatory hydrogenase HupUV of Rhodobacter capsulatus O2 sensitive without affecting its transductory activity. FEBS J. 272, 3899–3908 (2005)
Shomura, Y., Hagiya, K., Yoon, K.-S., Nishihara, H. & Higuchi, Y. Crystallization and preliminary X-ray diffraction analysis of membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus. Acta Crystallogr. F 67, 827–829 (2011)
Ueno, G., Kanda, H., Kumasaka, T. & Yamamoto, M. Beamline Scheduling Software: administration software for automatic operation of the RIKEN structural genomics beamlines at SPring-8. J. Synchrotron Radiat. 12, 380–384 (2005)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, Number 4 . The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Acknowledgements
We thank K. Hagiya for technical assistance at Ibaraki University. The synchrotron radiation experiments were performed at the BL41XU (proposal no. 2010A1223) and BL44XU (proposal no. 2010A/B6520) with the approval of JASRI. The CCD detector MX225-HE (Rayonix) at BL44XU was financially supported by Academia Sinica and National Synchrotron Radiation Research Center (Taiwan). This work was supported by a grant-in-aid for Scientific Research from MEXT (20051022 (Y.S.), 22770111 (Y.S.), 18GS0207 (Y.H.)), grant-in-aid for Scientific Research from JSPS (20580094 (H.N.), 22370061 (Y.H.) and 22657031 (Y.H.)), grant-in-aid for research and education from University of Hyogo (Y.S.), and grant-in-aid for young scientists from Hyogo Science and Technology Association (Y.S.). This work was also partially supported by the GCOE Program (Y.H.), the Japanese Aerospace Exploration Agency Project (Y.H.), and the basic research programs CREST type, ‘Development of the Foundation for Nano-Interface Technology’ from JST, Japan (Y.H.).
Author information
Authors and Affiliations
Contributions
K.-S.Y. and H.N. performed bacterial culture and protein purification under management by H.N. Y.S. performed crystallization, X-ray data collection and structure determination. Y.S. and Y.H. prepared the manuscript with contributions from all authors. The overall project management was done by Y.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-4 with legends, Supplementary Tables 1-3 and an additional reference. (PDF 2062 kb)
Rights and permissions
About this article
Cite this article
Shomura, Y., Yoon, KS., Nishihara, H. et al. Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature 479, 253–256 (2011). https://doi.org/10.1038/nature10504
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10504
This article is cited by
-
Distinction between film loss and enzyme inactivation in protein-film voltammetry: a theoretical study in cyclic staircase voltammetry
Monatshefte für Chemie - Chemical Monthly (2023)
-
Microbial oxidation of atmospheric trace gases
Nature Reviews Microbiology (2022)
-
Design of a gold clustering site in an engineered apo-ferritin cage
Communications Chemistry (2022)
-
Aerobic hydrogen-oxidizing bacteria in soil: from cells to ecosystems
Reviews in Environmental Science and Bio/Technology (2022)
-
Chemosynthetic and photosynthetic bacteria contribute differentially to primary production across a steep desert aridity gradient
The ISME Journal (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.