Abstract
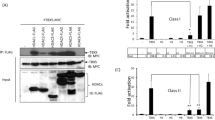
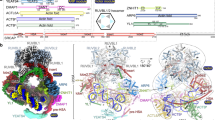
Histone lysine acetylation and methylation have an important role during gene transcription in a chromatin context1,2. Knowledge concerning the types of protein modules that can interact with acetyl-lysine has so far been limited to bromodomains1. Recently, a tandem plant homeodomain (PHD) finger3 (PHD1–PHD2, or PHD12) of human DPF3b, which functions in association with the BAF chromatin remodelling complex to initiate gene transcription during heart and muscle development, was reported to bind histones H3 and H4 in an acetylation-sensitive manner4, making it the first alternative to bromodomains for acetyl-lysine binding5. Here we report the structural mechanism of acetylated histone binding by the double PHD fingers of DPF3b. Our three-dimensional solution structures and biochemical analysis of DPF3b highlight the molecular basis of the integrated tandem PHD finger, which acts as one functional unit in the sequence-specific recognition of lysine-14-acetylated histone H3 (H3K14ac). Whereas the interaction with H3 is promoted by acetylation at lysine 14, it is inhibited by methylation at lysine 4, and these opposing influences are important during transcriptional activation of the mouse DPF3b target genes Pitx2 and Jmjd1c. Binding of this tandem protein module to chromatin can thus be regulated by different histone modifications during the initiation of gene transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
The structural coordinates of human DPF3b PHD12 in complex with H3K14ac, H3, H4-Nac, or H4K16ac peptides are deposited in the Protein Data Bank under accession codes 2kwj, 2kwk, 2kwn and 2kwo, respectively. Accession codes for the chemical shift assignments of the corresponding NMR structures deposited at the BioMagResBank are 16858, 16859, 16861 and 16865, respectively.
References
Sanchez, R. & Zhou, M. M. The role of human bromodomains in chromatin biology and gene transcription. Curr. Opin. Drug Discov. Devel. 12, 659–665 (2009)
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature Struct. Mol. Biol. 14, 1025–1040 (2007)
Bienz, M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 31, 35–40 (2006)
Lange, M. et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 22, 2370–2384 (2008)
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999)
Clore, G. M. & Gronenborn, A. M. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 239, 349–363 (1994)
Li, H. et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 (2006)
Peña, P. et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 (2006)
Ramón-Maiques, S. et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl Acad. Sci. USA 104, 18993–18998 (2007)
Matthews, A. G. et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450, 1106–1110 (2007)
Ooi, S. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007)
Chakravarty, S., Zeng, L. & Zhou, M. M. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure 17, 670–679 (2009)
Org, T. et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376 (2008)
Zeng, L., Zhang, Q., Gerona-Navarro, G., Moshkina, N. & Zhou, M. M. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 16, 643–652 (2008)
Wysocka, J. et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 (2006)
Fischle, W. et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005)
Ruthenburg, A. J., Li, H., Patel, D. J. & Allis, C. D. Multivalent engagement of chromatin modifications by linked binding modules. Nature Rev. Mol. Cell Biol. 8, 983–994 (2007)
Jacobson, R., Ladurner, A., King, D. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000)
Flanagan, J. et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438, 1181–1185 (2005)
Huang, Y., Fang, J., Bedford, M., Zhang, Y. & Xu, R. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312, 748–751 (2006)
Zeng, L. et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nature Struct. Mol. Biol. 15, 626–633 (2008)
Phanstiel, D. et al. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc. Natl Acad. Sci. USA 105, 4093–4098 (2008)
Mujtaba, S. et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell 13, 251–263 (2004)
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Rieping, W. et al. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23, 381–382 (2007)
Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996)
Portiér, G. L., Benders, A. G., Oosterhof, A., Veerkamp, J. H. & van Kuppevelt, T. H. Differentiation markers of mouse C2C12 and rat L6 myogenic cell lines and the effect of the differentiation medium. In Vitro Cell. Dev. Biol. Anim. 35, 219–227 (1999)
Furlan-Magaril, M., Rincon-Arano, H. & Recillas-Targa, F. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol. Biol. 543, 253–266 (2009)
Qian, C. et al. Structure and chromosomal DNA binding of the SWIRM domain. Nature Struct. Mol. Biol. 12, 1078–1085 (2005)
Acknowledgements
We acknowledge the use of the NMR Facility at the New York Structural Biology Center. We thank S. Hearn of Cold Spring Harbor Laboratory microscopy facility for help with confocal microscopy study, and A. Buku and G. Gerona-Navarro for technical advice on peptide binding analysis. This work was supported by grants from the National Institutes of Health (M.-M.Z.).
Author information
Authors and Affiliations
Contributions
M.-M.Z. and M.J.W. designed the study; Q.Z. generated constructs and protein samples, and contributed to the immunofluorescence study; L.Z. determined the protein structures; A.N.P. contributed to the protein biochemistry study; L.Z. and Q.Z. performed protein and peptide binding study; S.L. and M.J.W. carried out the cell biology experiments; M.-M.Z. supervised the project; all authors contributed to manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-12 with legends and Supplementary Tables 1-5. (PDF 8141 kb)
Rights and permissions
About this article
Cite this article
Zeng, L., Zhang, Q., Li, S. et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258–262 (2010). https://doi.org/10.1038/nature09139
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09139
This article is cited by
-
Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2
Nature Structural & Molecular Biology (2023)
-
Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1
Current Genetics (2023)
-
Identification of novel PHD-finger genes in pepper by genomic re-annotation and comparative analyses
BMC Plant Biology (2022)
-
Comprehensive characterization of three classes of Arabidopsis SWI/SNF chromatin remodelling complexes
Nature Plants (2022)
-
A natural allele of OsMS1 responds to temperature changes and confers thermosensitive genic male sterility
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.