Abstract
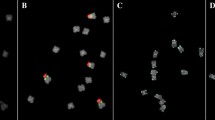
A modified DNA microarray-based technique was devised for preliminary screening of short fragment genomic DNA libraries from three Vicia species (V. melanops, V. narbonensis, and V. sativa) to isolate representative highly abundant DNA sequences that show different distribution patterns among related legume species. The microarrays were sequentially hybridized with labeled genomic DNAs of thirteen Vicia and seven other Fabaceae species and scored for hybridization signals of individual clones. The clones were then assigned to one of the following groups characterized by hybridization to: (1) all tested species, (2) most of the Vicia and Pisum species, (3) only a few Vicia species, and (4) preferentially a single Vicia species. Several clones from each group, 65 in total, were sequenced. All Group I clones were identified as rDNA genes or fragments of chloroplast genome, whereas the majority of Group II clones showed significant homologies to retroelement sequences. Clones in Groups III and IV contained novel dispersed repeats with copy numbers 102–106/1C and two genus-specific tandem repeats. One of these belongs to the VicTR-B repeat family, and the other clone (S12) contains an amplified portion of the rDNA intergenic spacer. In situ hybridization using V. sativa metaphase chromosomes revealed the presence of the S12 sequences not only within rDNA genes, but also at several additional loci. The newly identified repeats, as well as the retroelement-like sequences, were characterized with respect to their abundance within individual genomes. Correlations between the repeat distributions and the current taxonomic classification of these species are discussed.
Similar content being viewed by others
References
Alix, K., Baurens, F.C., Paulet, F., Glaszmann, J.C. and D'Hont, A. 1998. Isolation and characterization of a satellite DNA family in the Saccharum complex. Genome 41: 854–864.
Allkin, R., Goyder, D.J., Bisby, F.A. and White, R.J. 1986. Names and synonyms of species and subspecies in the Vicieae: Issue 3. Vicieae Database Project, Publication No 7.
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25: 3389–3402.
Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seid-man, J.G., Smith, J.A. and Struhl, K. 1991. Current Protocols in Molecular Biology. John Wiley, New York.
Bennett, M.D. and Leitch, I.J. 1998. Angiosperm DNA C-values database. http://www.rbgkew.org.uk/cval/database1.html.
Bennett, M.D. and Smith, J.B. 1976. Nuclear DNA amounts in angiosperms. Phil. Trans. R. Soc. Lond. B 274: 227–274.
Castellino, A.M. 1997. When the chips are down. Genome Res. 7: 943–946.
Chavanne, F., Zhang, D.X., Liaud, M.F. and Cerff, R. 1998. Structure and evolution of Cyclops: a novel giant retrotransposon of the Ty3/Gypsy family highly amplified in pea and other legume species. Plant Mol. Biol. 37: 363–375.
Chooi, W.Y. 1971. Comparison of the DNA of six Vicia species by the method of DNA-DNA hybridization. Genetics 68: 213–230.
Dellaporta, S.L., Wood, J. and Hicks, J.B. 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1: 19–21.
Dvořák, J. and Zhang, H.B. 1992. Reconstruction of the phylogeny of the genus Triticum from variation in repeated nucleotide-sequences. Theor. Appl. Genet. 84: 419–429.
Ellis, T.H.N., Poyser, S.J., Knox, M.R., Vershinin, A.V. and Ambrose, M.J. 1998. Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol. Gen. Genet. 260: 9–19.
Falquet, J., Creusot, F. and Dron, M. 1997. Molecular analysis of Phaseolus vulgaris rDNA unit and characterization of a satellite DNA homologous to IGS subrepeats. Plant Mol. Biochem. 35: 611–622.
Flavell, A.J., Pearce, S.R., Heslop-Harrison, J.S.P. and Kumar, A. 1997. The evolution of Ty1-copia group retrotransposons in eukaryote genomes. Genetica 100: 185–195.
Flavell, A.J., Smith, D.B. and Kumar, A. 1992. Extreme heterogenity of Ty1-copia group retrotransposons in plants. Mol. Gen. Genet. 231: 233–242.
Flavell, R.B., Bennett, M.D., Smith, J.B. and Smith, D.B. 1974. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet. 12: 257–269.
Frediani, M., Gelati, M.T., Maggini, F., Galasso, I., Minelli, S., Ceccarelli, M. and Cionini, P.G. 1999. A family of dispersed repeats in the genome of Vicia faba: structure, chromosomal organization, redundancy modulation, and evolution. Chromosoma 108: 317–324.
Grandbastien, M.A., Audeon, C., Casacuberta, J.M., Grappin, P., Lucas, H., Moreau, C. and Pouteau, S. 1994. Functional analysis of the tobacco Tnt1 retrotransposon. Genetica 93: 181–189.
Gualberti, G., Doležel, J., Macas, J. and Lucretti, S. 1996. Preparation of pea (Pisum sativum L.) chromosome and nucleus suspensions from single root tips. Theor. Appl. Genet. 92: 744–751.
Gwynne, P. and Page, G. 1999. Microarray analysis: the next revolution in molecular biology. Science 285: 911–938.
Kato, A., Iida, Y., Yakura, K. and Tanifuji, S. 1985. Sequence analysis of Vicia faba highly repeated DNA: the BamHI repeated sequence. Plant Mol. Biol. 5: 41–53.
Kehoe, D.M., Villand, P. and Somerville, S. 1999. DNA microarrays for studies of higher plants and other photosynthetic organisms. Trends Plant Sci. 4: 38–41.
Kidwell, M.G. and Lisch, D. 1997. Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA 94: 7704–7711.
King, K., Jobst, J. and Hemleben, V. 1995. Differential homogenization and amplification of two satellite DNAs in the genus Cucurbita (Cucurbitaceae). J. Mol. Evol. 41: 996–1005.
Kupicha, F.K. 1976. The infrageneric structure of Vicia.Notes R. Bot. Gard. Edinb. 34: 287–326.
Lehmann, P. and Kozubek, E. 1993. Genomic organization and nucleotide sequence of the repetitive DNA from Vicia sativa L. Genet. Pol. 34: 185–195.
Macas, J., Nouzová, M. and Galbraith, D.W. 1998. Adapting the Biomekr 2000 laboratory automation workstation for printing DNA microarrays. Biotechniques 25: 106–106.
Macas, J., Požárková, D., Navrátilová, A., Nouzová, M. and Neu-mann, P. 2000. Two new families of tandem repeats isolated from genus Vicia using genomic self-priming PCR. Mol. Gen. Genet. 263: 741–751.
Maggini, F., Cremonini, R., Zolfino, C., Tucci, G. F., D'Ovidio, R., Delre, V., De Pace, P.G., Scarascia Mugnozza, G.T. and Cionini, P.G. 1991. Structure and chromosomal localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma 100: 229–234.
Maggini, F., D'Ovidio, R., Gelati, M.T., Frediani, M., Cremonini, R., Ceccarelli, M., Minelli, S. and Cionini, P.. 1995. Fok I DNA repeats in the genome of Vici a faba species: specificity, structure, redundancy modulation, and nuclear organization. Genome 38: 1255–1261.
Marillonnet, S. and Wessler, S.R. 1998. Extreme structural heterogeneity among the members of a maize retrotransposon family. Genetics 150: 1245–1256.
Marschalek, R., Hofmann, J., Schumann, G., Gösseringer, R. and Dingermann, T. 1992. Structure of DRE, a retrotransposable element which integrates with position specificity upstream of Dictyostelium discoideum tRNA genes. Mol. Cell Biol. 12: 229–239.
Maxted, N. 1995. An ecogeographical study of Vicia subgenus Vicia. Systematic and ecogeographic studies on crop genepools. 8. International Plant Genetic Resources Institute, Rome.
Nakajima, R., Noma, K., Ohtsubo, E. and Ohtsubo, H. 1996. Identification and characterization of two tandem repeat sequences (TrsB and TrsC) and a retrotransposon (RIRE1) as genomegeneral sequences in rice. Genes Genet. Syst. 71:373–382.
Nouzová, M., Kubaláková, M., Doleželová, M.D., Koblížková, A., Neumann, P., Doležel, J. and Macas, J. 1999. Cloning and characterization of new repetitive sequences in field bean (Vicia faba L.). Ann. Bot. 83: 535–541.
Pearce, S.R., Harrison, G., Li, D.T., Heslop-Harrison, J.S., Kumar, A. and Flavell, A.J. 1996. The Ty1-copia group retrotransposons in Vicia species: copy number, sequence heterogeneity and chromosomal localization. Mol. Gen. Genet. 250: 305–315.
Pearson, W.R. and Lipman, D.J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85: 2444–2448.
Phimister, B. 1999. A note on nomenclature. Nature Genet. 21S: 1.
Polhill, R.M. and Raven, P.H. 1981. Papilionoideae. In: R.M. Polhill and P.H. Raven (Eds) Advances in Legume Systematics, Royal Botanic Gardens, Kew, UK, pp. 191–409
Potokina, E., Tomooka, N., Vaughan, D.A., Alexandrova, T. and Xu, R.Q. 1999. Phylogeny of Vicia subgenus Vicia (Fabaceae) based on analysis of RAPDs and RFLP of PCR-amplified chloroplast genes. Genet. Res. Crop Evol. 46: 149–161.
Raina, S.N. and Narayan, R.K.J. 1984. Changes in DNA composition in the evolution of Vicia species. Theor. Appl. Genet. 68: 187–192.
Raina, S.N. and Ogihara, Y. 1995. Ribosomal DNA repeat unit polymorphism in 49 Vicia species. Theor. Appl. Genet. 90: 477–486.
Sanger, F., Nicklen, D. and Coulson, A.R. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74: 5463–5467.
SanMiguel, P. and Bennetzen, J.L. 1998. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82: 37–44.
SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y. and Bennet-zen, J.L. 1998. The paleontology of intergene retrotransposons of maize. Nature Genet. 20: 43–45.
Schena, M., Shalon, D., Davis, R.W. and Brown, P.O. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470.
Schena, M., Shalon, D., Heller, R., Chai, A., Brown, P.O. and Davis, R.W. 1996. Parallel human genome analyses: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93: 10614–10619.
Schmidt, T. and Heslop-Harrison, J.S. 1993. Variability and evolution of highly repeated DNA-sequences in the genus Beta. Genome 36: 1074–1079.
Schmidt, T. and Heslop-Harrison, J.S. 1998. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends Plant Sci. 3: 195–199.
Shalon, D., Smith, S.J. and Brown, P.O. 1996. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 6: 639–645.
Solano, R., Hueros, G., Fominaya, A. and Ferrer, E. 1992. Organization of repeated sequences in species of the genus Avena. Theor. Appl. Genet. 83: 602–607.
Staginnus, C., Winter, P., Desel, C., Schmidt, T. and Kahl, G. 1999. Molecular structure and chromosomal localization of major repetitive DNA families in the chickpea (Cicer arietinum L.) genome. Plant Mol. Biol. 39: 1037–1050.
Suoniemi, A., Schmidt, D. and Schulman, A.H. 1997. BARE-1 insertion site preferences and evolutionary conservation of RNA and cDNA processing sites. Genetica 100: 219–230.
Suoniemi, A., Tanskanen, J. and Schulman, A.H. 1998. Gypsy-like retrotransposons are widespread in the plant kingdom. Plant J. 13: 699–705.
Svitashev, S., Bryngelsson, T., Vershinin, A., Pedersen, C., Sall, T. and Vonbothmer, R. 1994. Phylogenetic analysis of the genus Hordeum using repetitive DNA sequences. Theor. Appl. Genet. 89: 801–810.
Unfried, K., Schiebel, K. and Hemleben, V. 1991. Subrepeats of rDNA intergenic spacer present as prominent independent satellite DNA in Vigna radiata but not in Vigna angularis. Gene 99: 63–68.
Vogt, P. 1992. Code domains in tandem repetitive DNA-sequence structures. Chromosoma 101: 585–589.
Voytas, D.F., Cummings, M.P., Konieczny, A.K., Ausubel, F.M. and Rodermel, S.R. 1992. Copia-like retrotransposons are ubiquitous among plants. Proc. Natl. Acad. Sci USA 89: 7124–7128.
Yakura, K. and Tanifuji, S. 1983. Molecular cloning and restriction analysis of EcoRI fragments of Vicia faba rDNA. Plant Cell Physiol. 24: 1327–1330.
Zhao, X. and Kochert, G. 1993. Clusters of interspersed repeated DNA sequences in the rice genome (Oryza). Genome 36: 944–953.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nouzová, M., Neumann, P., Navrátilová, A. et al. Microarray-based survey of repetitive genomic sequences in Vicia spp.. Plant Mol Biol 45, 229–244 (2001). https://doi.org/10.1023/A:1006408119740
Issue Date:
DOI: https://doi.org/10.1023/A:1006408119740