Abstract
SARS-CoV-2 is the virus responsible for the COVID-19 pandemic, and its effects on people worldwide continue to grow. Protein-targeted therapeutics are currently unavailable for this virus. As with other coronaviruses, the nucleocapsid (N) protein is the most conserved RNA-binding structural protein of SARS-CoV-2. The N protein is an appealing target because of its functional role in viral transcription and replication. Therefore, molecular docking method for structure-based drug design was used to investigate the binding energy and binding modes of various anti-N inhibitors in depth. The inhibitors selected were originally developed to target stress granules and other molecules involved in RNA biology, and were either FDA-approved or in the process of clinical trials for COVID-19. We aimed at targeting the N-terminal RNA binding domain (NTD) for molecular docking-based screening, on the basis of the first resolved crystal structure of SARS-CoV-2 N protein (PDB ID: 6M3M) and C-terminal domain (CTD) dimerization of the nucleocapsid phosphoprotein of SARS-COV-2 (PDB ID: 6WJI). Silmitasertib, nintedanib, ternatin, luteolin, and fedratinib were found to interact with RNA binding sites and to form a predicted protein interface with high binding energy. Similarly, silmitasertib, sirolimus-rapamycin, dovitinib, nintedanib, and fedratinib were found to interact with the SARS-CoV-2 N protein at its CTD dimerization sites, according to previous studies. In addition, we investigated an information gap regarding the relationships among the energetic landscape and stability and drug binding of the SARS-CoV-2 N NTD and CTD. Our in silico results clearly indicated that several tested drugs as potent putative inhibitors for COVID-19 therapeutics, thus indicating that they should be further validated as treatments to slow the spread of SARS-CoV-2.
Similar content being viewed by others
1 Introduction
Since the emergence of the Delta variant of SARS-CoV-2 in May 2021, more than 155 million medically confirmed cases and approximately 3.24 million deaths occurred in more than 180 countries as a result of the COVID-19 pandemic [1]. The SARS-CoV-2/2019-nCoV novel coronavirus strain causes this respiratory disease [2, 3]. Repurposing of drugs has been a promising strategy at the forefront of strategies to address the continually growing number of COVID-19 cases [4,5,6,7,8]. In the current pandemic situation, drug repurposing should be considered a new avenue for the treatment of COVID-19 [5]. The coronavirus virion consists of structural proteins, namely spike (S), envelope (E), membrane (M), nucleocapsid (N) and, for some beta coronaviruses, hemagglutinin esterase [9]. The genomic structure of SARS-CoV-2 contains short untranslated regions and N-3′, E, M, 5′-replicase (rep)-S, identical to the genome structures of other coronaviruses [10]. As in other coronaviruses, the N protein is a crucial structural component of SARS-CoV-2 [11]. SARS-CoV-2 N has 90% sequence identity to the Severe Acute Respiratory Syndrome coronavirus N protein [12].
The N protein, which is naturally found within the virus, is the most conserved structural protein. It is required for viral replication and transcription, as it binds the viral RNA genome [13]. The N protein structure comprises an N-terminal RNA-binding domain (NTD), a C-terminal domain (CTD), and a naturally disordered central Arg/Ser-rich linker. For the SARS-CoV-2 N protein, each NTD molecule adopts a right-handed fist shape. The core sub domain consists of a five-stranded U-shaped antiparallel β-sheet with β4–β2–β3–β1–β5 topology, sandwiched between two short α-helices (α1 before the β2 strand and α2 after the β5 strand), and a protruding β-hairpin (β2′–β3′) is composed of mostly basic amino acid residues. New COVID-19 drug targets have been identified in the SARS-CoV-2-human interactome recently published by Gordon et al. [14]. Virtual screening and molecular docking have led to the identification of various inhibitors of SARS-CoV-2 [15].
Among the 332 interactions previously identified between viral and host proteins, most involve the innate immune signaling pathway [13, 16]. With this knowledge, researchers have identified an anti-N drug chain that shows excellent promise as a treatment for COVID-19 [17]. Through pathway analysis, a series of anti-N drugs with high potential to combat COVID-19 have been identified. Interestingly, some of the drugs target the N protein, which has been suggested to be a viable target for antiviral drug development [18]. The viral N protein targets the mTOR translational repressors LARP1, protein kinase CK2, and stress granule protein G3BP1/2. Viral replication is inhibited by stress granules and the host translation machinery [19, 20], whereas viruses suppress stress granules and use the host’s translation machinery for viral replication. Because of their roles in RNA genomic packaging, viral transcription, and replication, several recent studies have shown that coronavirus N proteins can be valuable antiviral drug targets [21,22,23,24,25]. Recently, studies have shown that the N proteins of coronavirus are valuable antiviral drug targets, owing to their roles in viral transcription, RNA genomic packing, and replication [26]. As C-terminal domain of N protein (CTD) is involved in the self-assembly of N protein into a filament that is packaged into new virions, so structure-based molecular docking experiments have been performed on the CTD for the identification of possible inhibitors of N protein [17].
The first available crystal structures of N protein NTD (PDB ID:6M3M), abbreviated as N-NTD in this manuscript, and CTD (PDB ID: 6WJI) were published in late April 2020 [27]. Herein, these X-ray crystal structures were retrieved from the Protein Data Bank to conduct molecular docking to test the drugs recently described by Gordon et al. We considered all six FDA-approved drugs on the list, five drugs in clinical trials, and other compounds from the IUPHAR/BPS Guide to Pharmacology (2020-3-12) in our studies. This work focused on the identification of high confidence candidate drugs, and their effective roles in therapeutic interventions against COVID-19.
2 Methods
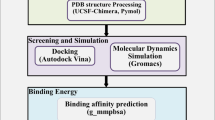
2.1 Molecular Docking
From the Protein Data Bank of RCSB (https://www.rcsb.org), we retrieved three-dimensional crystal structures of the SARS-CoV-2 N-NTD (PDB ID: 6M3M) and C-CTD (PDB ID: 6WJI), which were used as models for molecular docking analysis. PyRx, AutoDock Vina [28], and AutoDock Tools [29] were used for molecular docking analysis. SARS-CoV-2 C-CTD and N-NTD pockets at the binding site were selected with AutoDock Tools to create the grid box boundaries for docking [29]. For ligand preparation, such as charges, root detection, aromaticity, and hydrogen, the protein structure was converted to.pdbqt format (an accepted format for AutoDock Vina). Protein preparation for N-NTD and C-CTD proteins was performed by first adding the missing hydrogen atoms, then removing water and any other metal ions. PubChem (https://pubchem.ncbi.nlm.nih.gov) was used as a source to retrieve the structures of drugs. In PyRx, we converted all drug molecules from PDB to PDBQTs [29]. We took advantage of using PyRX rather than Vina because it can dock many more compounds in one session. The structure of proteins was pre-processed in PyMOL [30] by deletion of the chains, unoccupied binding sites, and ligands/water molecules from the protein structure. For docking purposes, the protein preparation was performed by addition of hydrogens and charges with the AutoDock tool. AutoDock Vina was used for molecular docking. The docking conformations of the top pose were obtained, and energy minimization of post-docking was performed in Discovery Studio (DS 3.531). Then PyMOL [30], Ligplot+ [31], and Discovery studio visualizer (Dassault Systèms BIOVIA 2017) were used to visualize and study the resultant receptor-ligand docked complexes.
2.2 Drug–Target Network Construction
We used the STITCH (Search Tool for Interactions of Chemicals; www.stitch.embl.de) web server [32] to rationally select possible drug targets for drug–target network construction. The targets were identified on the basis of a network interaction score above 0.9.
3 Results
3.1 Binding Site Exploration
The CASTp Server was used to predict the binding sites by searching for CTD ligand-independent binding sites to perform molecular docking [33]. The CASTp Server identified a putative binding pocket with a volume of 1166 Å3 and a surface area of 907 Å2 (solvent accessible). The CTD binding pocket consisted of a coil region (335–349) at the C-terminus, and β2 (329–334), 259–264 (310 helix), H4 (310–311, 314–318), 270, 274 (helix H1), H3 (301, 304–306), H2 (291, 292, 295–296), and 281–287 (turn and coil region between H1 and H5) residues (Supplementary Fig. S1). To predict the binding sites, we used two additional web servers, PrankWeb [34] and COACH [35]. Similar binding pocket residues with minor differences were also predicted with these two servers. Interestingly, both Protein–Protein Interaction Site Predictor (cons-PPISP) server and InterProSurf analysis indicated that most of the binding pocket residues were predicted to be potential dimeric interfacial residues [36, 37]. The results from these two web-servers reflected that the CTD dimerization residues are located in the coil regions adjacent to β1 (Arg319–Val324), β2 (Gly335–Leu339), H4 (Ser310–Ser318), and β2 (Gly328–Thr334).
3.2 Molecular Docking
PPIs of SARS-CoV-2-human with 332 high-confidence and 69 candidate drugs have been identified to potentially treat COVID-19 in recent research performed by the Quantitative Biosciences Institute of University of California San Francisco.
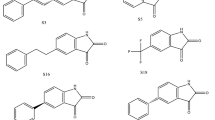
Some of these drugs target the N protein and are already on the market, whereas others remain in phase 3 clinical trials. To verify the binding efficacy of compounds against the targets, we performed molecular docking analysis on the N-NTD (PDB ID: 6M3M) and C-CTD (PDB ID: 6WJI) proteins. On the basis of the binding affinity in kcal/mol, 20 potential drugs for COVID-19 were considered (Fig. 1). These drugs work in a variety of ways and include CK-2 inhibitors, mTOR inhibitors, translation inhibitors, SG inhibitors, and multi-targeted protein kinase inhibitors.
3.3 Molecular Interactions Between the SARS-CoV-2 N-NTD and Drugs
Virtual screening of compounds was followed by docking of potential high confidence drugs as well as candidate drugs, and the results were evaluated on the basis of binding affinity. Each of the 20 drugs were docked against the and evaluated according to their binding affinity against SARS-CoV-2 proteins (Table 1). Notably, the key residues involved in the RNA binding interactions, including S51, F53, R107, Y109, Y111, and R149 (in SARS-CoV-2 N-NTD numbering), are conserved, thus suggesting their potential for drug development. According to their binding affinities and the visual inspection of docked complexes for their ability to form hydrogen bonds and other interactions with the N-NTD, the best five molecules were chosen (Fig. 2). The compounds (and their corresponding binding affinities) were as follows: fedratinib (− 7.3 kcal/mol), luteolin (− 7.5 kcal/mol), nintedanib (− 8.2 kcal/mol), and ternatin (− 7.8 kcal/mol). Among all the compounds, silmitasertib showed the highest binding affinity (− 8.8 kcal/mol), exceeding that of the approved drug remdesivir (− 5.79 kcal/mol; [7]). Fedratinib bound with the least binding affinity (− 7.3 kcal/mol). Moreover, foretinib, dovitinib, axitinib, staurosporine, and sirolimus also exhibited strong binding, with binding energies of − 7.0 kcal/mol. As shown in Fig. 2A, silmitasertib, a casein kinase-2 inhibitor, showed the highest binding affinity with hydrogen-bonding residue Thr77, and hydrophobic interactions with Trp53, Ile75, Ile147, and Ile158, along with several van der Waals interactions with Asn76, Asn78, Asn155, and Ala156. Nintedanib, a tyrosine kinase inhibitor, formed two hydrogen bonds with Thr77, while the residues such as Trp53, Ala153, Ile158, and Val159 interacted hydrophobically, along with van der Waals interactions with three Asn residues (76, 78, and 155) (Fig. 2B).
Molecular docking interactions and orientations of the top five anti-N drugs with the SARS-CoV-2 N-NTD. Docking interactions of A silmitasertib, B nintedanib, and C ternatin. The right panel shows a schematic representation of interactions of these drug molecules with the N-NTD. The protein residues and interactions are colored accordingly and provided in the figure
Ternatin, a protein biogenesis inhibitor, formed five hydrogen bonds with Gly70, Gln84, Thr136, Gln164, and Thr166, along with hydrophobic interactions with residues including Val73, Ile75, Pro81, and Pro163 (Fig. 2C). Luteolin, an antiviral flavone, displayed hydrogen bonds involving Gln59, Glu63, Asp64, Arg90, and Arg93, along with hydrophobic and van der Waals interactions with Leu65, Thr92, Arg94, Leu105, Pro107, Trp109, and Lys170, displaying strong binding affinity (Fig. 3A).
Molecular docking interactions and orientations of the top five anti-N drugs with the SARS-CoV-2 N-NTD. Docking interactions of A luteolin and B fedratinib. The right panel shows a schematic representation of interactions of these drug molecules with the N-NTD. The protein residues and interactions are colored accordingly and provided in the figure
The JAK2 inhibitor fedratinib interacts with the N-NTD through hydrogen bonds with Lys66 and Thr58, along the with hydrophobic interactions mediated by Ala51, Ser52, Thr55, His60, Ala91, Tyr110, Ala157, and Ala174. The binding is also characterized by van der Waals interactions with Thr55, Arg93, Arg108, and Arg150 (Fig. 3B). The fedratinib binding sites are similar to the RNA binding sites, thus suggesting that fedratinib binding may hinder the RNA binding of N protein.
Another anticancer drug, foretinib, interacted with the N-NTD via two hydrogen bonds involving Asn76, Ile147, and Gln161. Other key residues interacting with foretinib were Trp53, Ile75, Thr77, Asn78, Tyr113, His146, Gly148, and Ile158 (Supplementary Fig. S1). The drugs dovitinib, staurosporine, and zotatifin also showed considerable binding affinity and interacted thorough hydrogen bonds, hydrophobic interactions, and van der Waals interactions (Supplementary Fig. S2).
Hence, the hydrogen bonding and hydrophobic interactions were the driving forces for binding, as indicated by the binding modes of top ranked drugs. The key residues of the N-NTD interacting with the drugs were Trp53, Ile75, Asn76, Thr77, Arg93, Arg94, Ala153, Ala156, Ile158, Val159, and Gln161, as shown by the results obtained from all drug docking calculations (Table 1). These residues are largely found in the turn region and are involved in ribonucleotide binding and protein–protein interfaces. Through the results of the cons-PPISP server, and InterProSurf analysis, residues in the β3 strand (Thr92–Arg96), the turn residues between β5 and β6 (Gly117, Pro118, Ala120, and Gly121), and the C-terminal tail (Asn151, Asn154, Ala157, Val159, and Gln161) were predicted to be potential PPI interfaces.
3.4 Molecular Interactions Between the SARS-CoV-2 C-CTD and Drugs
All 20 drugs were docked against the C-CTD of N protein and rated according to their binding efficiency (Table 2). On the basis of their binding affinities and visual inspection of docked complexes for their ability to influence hydrogen bonds and other interactions with the C-CTD, the best five molecules were chosen (Fig. 4). The best five molecules (and their binding affinities) were fedratinib (− 8.2 kcal/mol), nintedanib (− 8.4 kcal/mol), sirolimus-rapamycin (− 9.3 kcal/mol), and dovitinib (− 8.6 kcal/mol); among these, silmitasertib showed the greatest binding affinity (− 9.3 kcal/mol).
Molecular docking interactions and orientations of the top five anti-N drugs with the SARS-CoV-2 C-CTD. Docking interactions of A silmitasertib, B sirolimus-rapamycin, and C dovitinib. The right panel shows a schematic representation of interactions of these drug molecules with the C-CTD. The protein residues and interactions are colored accordingly and provided in the figure
As shown in Fig. 4A, silmitasertib, a casein kinase-2 inhibitor, showed the highest binding affinity and formation of hydrogen bonds with Glu281, and Ser327, and hydrophobic interactions with Thr282, Ala336, Arg259, Ala336, Thr334, Gly335, Thr334, Thr332, Thr325, and Pro326, along with several van der Waals interactions.
As shown in Fig. 4B, rapamycin (sirolimus), a potent immunosuppressive agent, blocks or inhibits cytokine-mediated signal transduction pathways during late T-cell cycle progression. This inhibition occurs through modulation of the function of a target protein. Rapamycin forms hydrogen bonds with Asn354 and hydrophobic interactions with Ile357, Lys361, Asp362, Ala359, Lys355, Ile351, Val350, Val324, Met322, and Glu323, along with several van der Waals interactions. Figure 4C shows that dovitinib is as anticancer drug that inhibits multiple kinases. It forms hydrogen bonds with the C-CTD through AspB:840, LysB:338, and GlnA:260. Dovitinib binds the C-CTD of N through hydrophobic and van der Waals interactions with Asp343, Pro258, Lys257, Arg259, Arg262, Ile337, Leu339, and Phe307. Nintedanib, a tyrosine kinase inhibitor, forms two hydrogen bonds with Arg262 and hydrophobic interactions with Gly275, Phe274, Gln283, Thr262, Asp343, Asp340, Pro258, Gln349, Phe307, Ile337, Gln260, Leu339, Arg259, and Trp330, along with van der Waals interactions with three Asn residues (76, 78, and 155) (Fig. 5A). The JAK2 inhibitor fedratinib interacts with residues such as Asp340 and Asp343 via hydrogen bonding, and forms hydrophobic interactions mediated by Gly275, Arg276, Gly284, Gly283, Thr282, Phe274, Lys338, Trp330, Arg259, Gln349, Gln306, Gln260, Pro258, and Leu339 (Fig. 5B).
Molecular docking interactions and orientations of the top five anti-N drugs with the SARS-CoV-2 C-CTD. Docking interactions of A nintedanib and B fedratinib. The right panel shows a schematic representation of interactions of these drug molecules with the C-CTD. The protein residues and interactions are colored accordingly and provided in the figure
3.5 Interactions Between SARS-CoV-2 N and Human Proteins
Drug targets for drug repurposing were predicted by mapping the protein–protein interactions of SARS-CoV-2-human [14, 16]. Yellow color indicates human drug target interaction with SARS-CoV-2 (Fig. 6A). G3BP1, G3BP2, and LARP1 human proteins interact with N of SARS CoV-2 and are drug response proteins. Host proteins are involved in RNA splicing, viral defense, ribosome biogenesis in eukaryotes, metabolism of RNA, and mRNA catabolic processes (Fig. 6B).
3.6 Identification of Potential Drug–Target Interactions
The STITCH database was used as a model for drug–protein interactions through a statistical approach. STITCH incorporates data from 2031 genomes on more than 5 million interactions between 430,000 chemicals and 9.6 million proteins. To predict protein–drug interactions, it primarily relies on keyword mining of the literature and experimental evidence. The likelihood that the expected relationship occurs is indicated by a confidence score (0–0.9). A confidence score of 0.9 or higher was used to discover the targets. To construct a network based on binding affinities (\({K}_{i}\) of protein–drug interactions with thickness of edges between nodes, increasing as \({K}_{i}\) value increases), we used STITCH, which is based on STRING v10 [38]. Figure 7 shows the STITCH predictions for the drug–gene relationships among the top five strongly binding drugs. Some of these drugs had relatively fewer high affinity binding targets. Prominent proteins included YES1, RET, FGR, and FGFR1/2/3, which are involved in cancer and associated pathways as well as endocytosis, and dovitinib has been found to target some of these proteins (Fig. 7A). Fedratinib interacted with the JAK2/JAK1 proteins, which are part of the JAK-STAT signaling pathway (Fig. 7B).
Luteolin, a flavone, had a low affinity for the proteins CASP3, JUN, CDK2, FOS, and MAPK8, which are involved in the TNF signaling pathway and cancer (Fig. 7C). However, certain drugs had several predicted protein interactions and interacted via various pathways. For example, nintedanib targets the Ras signaling pathway, cancer, and cytokine–cytokine receptor interaction proteins MAP3K7, JAK1, PDGFRA/B, LCK, KIT, MELK, FLT3, and KDR (Fig. 7D). Similarly, sirolimus/rapamycin, a kinase inhibitor, interacts with many targets in the mTOR and AMPK signaling pathways, and with proteins involved in immunosuppression, including mTOR, FKBP1A, and FKBP5 (Fig. 7E). Silmitasertib interacts with CSNK2A1, a serine/threonine kinase protein involved in cell cycle progression, apoptosis, transcription, and viral infection (Fig. 7A). Another drug, foretinib, is involved in endocytosis and focal adhesions, and has similar targets to nintedanib and dovitinib.
4 Discussion
As the RNA binding activity of N protein is essential for viral ribonucleoprotein formation and genome replication, blocking the RNA binding of the N-NTD has been demonstrated to be a potential treatment strategy. N protein is an essential RNA-binding protein with crucial roles in replication and transcription of viral RNA. An overall right-handed fold with a β-sheet core is found between loops, as revealed by recently solved crystal structures of the SARS-CoV-2 N-NTD (PDB ID: 6M3M) and C-CTD (PDB ID: 6WJI). The core region of the β-sheet consists of five antiparallel β-strands with a β6–β2–β5–β1–β7 topology flanked by a single short α-helix just before strand β2, and a protruding β-hairpin (β3 and β4) between strands β2 and β5. In addition, an NMR structure of the SARS-CoV-2 N-NTD in complex with RNA (PDB ID: 6YI3) suggested putative RNA binding sites of A51, T58, H60, R93, I95, L105, S106, R108, R150, and Y173. Moreover, the adenosine monophosphate (AMP) binding site has been structurally characterized in HCoV-OC43 N-NTD by Lin et al. [22]. N49, A51, S52, A56, R89, R108, Y110, Y112, and R150 compose the AMP binding site, as indicated by the structural superposition between the SARS-CoV-2 N-NTD and HCoV-OC43 N-NTD-AMP. Therefore, we extended our investigation by using structure-based molecular docking of N-NTD with different drugs to gain insights into the structural and molecular regions’ potential effectiveness in antiviral drug therapy. The investigated drugs included protein biogenesis inhibitors, anticancer compounds, antiinflammatory compounds, mTOR inhibitors, and stress granule modifiers. The docking results showed that 5 of the 20 drugs bound with strong binding affinity. Among N-NTD inhibitors, the key residues involved in binding were near the helix, i.e., Trp53, Ile75, Asn76, and Thr77; in the β3 strand, i.e., Arg93 and Arg94; and at the C-terminal interface, i.e., Ala153, Ala156, Ile158, Val159, and Gln161.
Silmitasertib is an antiviral drug that has been tested against the N protein of SARS-CoV-2, and found to block the CK2 and enhance SGs formation [39], thus inhibiting SARS-CoV-2 proliferation in vitro. Recently, Taiwan-headquartered Senhwa Biosciences Inc and the US National Institutes of Health (NIH) collaborated in analyzing the effectiveness of silmitasertib for the treatment of COVID-19 (https://www.biospectrumasia.com/news/34/15848/senhwa-biosciences-nih-to-co-develop-COVID-19-drug.html). The drug showed promise in controlling the proliferation of this RNA virus in human clinical tests. Silmitasertib was developed by Senhwa Biosciences to treat cancers, such as pediatric brain tumors, medulloblastoma, and bile duct cancer.
The second molecule tested was nintedanib, a tyrosine kinase inhibitor used to treat idiopathic pulmonary fibrosis or interstitial lung disease [40]. Very recently, the safety and efficacy of nintedanib ethanesulfonate have been analyzed in treating pulmonary fibrosis in patients with mild-to-extreme COVID-19. A placebo-controlled, a single-center, randomized study has been initiated and is currently in a phase 2 clinical trial (ClinicalTrials.gov identifier: NCT04338802).
In addition, the viral translation inhibitors ternatin and zotatifin, which is an FDA-approved drug for the treatment of multiple myeloma, have demonstrated the strongest binding affinity [41]. Plitidepsin is structurally similar to ternatin and is currently undergoing a clinical trial in COVID-19. The flavone luteolin, an antiinflammatory molecule, has broad antiviral properties [42, 43]. Previous studies have shown that luteolin inhibits SARS-CoV S protein and 3CL protease [44, 45]. Recently, both luteolin and quercetin have been identified through virtual screening and molecular docking as the best possible SARS-CoV-2 inhibitors [46, 47]. Furthermore, through SUMMIT, the world's most powerful supercomputer, high-throughput screening of small molecules interacting with the SARS-CoV-2 S protein or S protein–human ACE2 interface have recently been reported. Eriodictyol, a structural analog of luteolin, has been found to be a potential inhibitor of SARS-CoV-2 [48]. The last drug with considerable binding affinity toward N protein is fedratinib, an antiinflammatory JAK2 inhibitor. Wu et al. [49] have reported that fedratinib suppresses the expression of IL17, IL 22, and L23 in murine TH17 cells, and suggested that the drug may help mitigate the cytokine storm associated with SARS-CoV-2 infection. Stebbing et al. [50], through in silico artificial intelligence, have predicted significant beneficial effects of the antiinflammatory agents baricitinib, fedratinib, and ruxolitinib in the treatment of COVID-19.
The drug foretinib is an anticancer agent that inhibits vascular endothelial growth factor receptor (VEGFR) and hepatocyte growth factor receptor (HEGFR or MET) receptor. A recent study has reported that foretinib (DB12307) is a strong binder, on the basis of analysis through in silico virtual screening and molecular docking of 8548 ligands on the SARS-CoV-2 endoribonuclease NendoU (PDB ID: 6VWW) [51]. Our findings also have suggested that this drug binds the nucleocapsid protein with high binding affinity. Thus, foretinib may be repurposed as a broad-spectrum drug and tested against COVID-19 in the future.
Intriguingly, the docking of the N-NTD and C-CTD proteins revealed promising results for all five drugs tested. Notably, fedratinib and luteolin bind the ribonucleotide binding site and thus can inhibit RNA binding of the protein. Similarly, silmitasertib and nintedanib are positioned at the interface of two monomers and thus can impair the oligomerization of the protein. We recommend further experimental investigation of these compounds.
5 Conclusion
The highly immunogenic and abundant nature of the N protein makes it a novel target to treat infection of the respiratory system by SARS-CoV2. We extended the investigation of drug efficacy, stimulated by the recent SARS-CoV-2-host interactome and identification of several anti-N drugs, by using computational analysis. In this study, binding modes were chosen, and the most common anti-N drugs were selected. The probable molecular underpinnings of their effectiveness against COVID-19 have been identified. The docking results indicated that 5 of 20 anti-N inhibitors bind with the energetic landscape of a protein–drug complex and have high thermodynamic scores. The identified drugs have been shown to bind the ribonucleotide binding pocket and protein interface of the N-NTD and C-CTD, thereby suggesting mechanisms of action. Thus, the identification of compounds that bind the N-NTD and C-CTD and interfere with NTD–RNA and NTD–NTD interactions may assist in the development of broad-spectrum antiviral therapeutics.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–54.
Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IA, Datir R, Collier DA, Albecka A, Singh S, Pandey R. SARS-CoV-2 B. 1.617. 2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–9.
Sanayaima Singh RK, Zubbair Malik M, Brojen Singh RK. Diversity of SARS-CoV-2 isolates driven by pressure and health index. Epidemiol Infect. 2021;149: e38.
Rosa SGV, Santos WC. Clinical trials on drug repositioning for COVID-19 treatment. Rev Panam Salud Publica. 2020;44: e40.
Gupta RK, Nwachuku EL, Zusman BE, Jha RM, Puccio AM. Drug repurposing for COVID-19 based on an integrative meta-analysis of SARS-CoV-2 induced gene signature in human airway epithelium. PLoS One. 2021;16(9): e0257784.
Gupta V, Haider S, Verma M, Singhvi N, Ponnusamy K, Zubbair Malik M, Verma H, Kumar R, Sood U, Hira P, Satija S, Singh Y, Lal R. Comparative genomics and integrated network approach unveiled undirected phylogeny patterns, co-mutational hotspots, functional crosstalk and regulatory interactions in SARS-CoV-2. mSystems. 2021;6: e0003021.
Mishra CB, Pandey P, Sharma RD, Malik MZ, Mongre RK, Lynn AM, Prasad R, Jeon R, Prakash A. Identifying the natural polyphenol catechin as a multitargeted agent against SARSCoV-2 for the plausible therapy of COVID-19: an integrated computational approach. Brief Bioinform. 2021;22:1346–60.
Alam A, Khan A, Imam N, Siddiqui MF, Waseem M, Malik MZ, Ishrat R. Design of an epitope-based peptide vaccine against the SARS-CoV-2: a vaccine-informatics approach. Brief Bioinform. 2021;22:1309–23.
V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–70.
Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, Kumar S, Bhattacharyya A, Kumar H, Bansal S, Medhi B. Drug targets for corona virus: a systematic review. Indian J Pharmacol. 2020;52(1):56–65.
Bai Z, Cao Y, Liu W, Li J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13(6):1115.
Martínez YA, Guo X, Portales-Pérez DP, Rivera G, Castañeda-Delgado JE, García-Pérez CA, Enciso-Moreno JA, Lara-Ramírez EE. The analysis on the human protein domain targets and host-like interacting motifs for the MERS-CoV and SARS-CoV/CoV-2 infers the molecular mimicry of coronavirus. PLoS One. 2021;16(2): e0246901.
Chang CK, Lo SC, Wang YS, Hou MH. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov Today. 2016;21(4):562–72.
Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–68.
Bhowmik D, Nandi R, Jagadeesan R, Kumar N, Prakash A, Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect Genet Evol. 2020;84: 104451.
Banaganapalli B, Al-Rayes N, Awan ZA, Alsulaimany FA, Alamri AS, Elango R, Malik MZ, Shaik NA. Multilevel systems biology analysis of lung transcriptomics data identifies key miRNAs and potential miRNA target genes for SARS-CoV-2 infection. Comput Biol Med. 2021;135:104570.
Yang M, He S, Chen X, Huang Z, Zhou Z, Zhou Z, Chen Q, Chen S, Kang S. Structural insight into the SARS-CoV-2 nucleocapsid protein C-terminal domain reveals a novel recognition mechanism for viral transcriptional regulatory sequences. Front Chem. 2021;8:1238.
Dai W, Zhang B, Jiang XM, Su H, Li J, Zhao Y, Xie X, Jin Z, Peng J, Liu F, Li C. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–5.
Nakagawa K, Narayanan K, Wada M, Makino S. Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol. 2018;92(20):e00902-e918.
Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol. 2019;11(5): a032813.
Chenavas S, Crepin T, Delmas B, Ruigrok RW, Slama-Schwok A. Influenza virus nucleoprotein: structure, RNA binding, oligomerization and antiviral drug target. Future Microbiol. 2013;8(12):1537–45.
Lin SY, Liu CL, Chang YM, Zhao J, Perlman S, Hou MH. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57(6):2247–57.
Lo YS, Lin SY, Wang SM, Wang CT, Chiu YL, Huang TH, Hou MH. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013;587(2):120–7.
Monod A, Swale C, Tarus B, Tissot A, Delmas B, Ruigrok RW, Crepin T, Slama-Schwok A. Learning from structure-based drug design and new antivirals targeting the ribonucleoprotein complex for the treatment of influenza. Expert Opin Drug Discov. 2015;10(4):345–71.
Sarma P, Sekhar N, Prajapat M, Avti P, Kaur H, Kumar S, Singh S, Kumar H, Prakash A, Dhibar DP, Medhi B. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). J Biomol Struct Dyn. 2020;2020:1–11.
Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat Rev Mol Cell Biol. 2022;23(1):21–39.
Kang S, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10(7):1228–38.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61.
Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–50.
Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24(5):417–22.
Laskowski RA, Swindells MB. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–86.
Szklarczyk D, Santos A, von Mering C, et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380-384.
Tian W, Chen C, Lei X, et al. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–7.
Jendele L, Krivak R, Skoda P, et al. PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019;47:W345–9.
Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–95.
Negi SS, Schein CH, Oezguen N, et al. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics. 2007;23:3397–9.
Chen H, Zhou HX. Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Proteins. 2005;61:21–35.
Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447-452.
Reineke LC, Tsai WC, Jain A, Kaelber JT, Jung SY, Lloyd RE. Casein kinase 2 is linked to stress granule dynamics through phosphorylation of the stress granule nucleating protein G3BP1. Mol Cell Biol. 2017;37(4):e00596-e616.
Flaherty KR, Wells AU, Cottin V, Devaraj A, Walsh SLF, Inoue Y, Richeldi L, Kolb M, Tetzlaff K, Stowasser S, Coeck C, Clerisme-Beaty E, Rosenstock B, Quaresma M, Haeufel T, Goeldner RG, Schlenker-Herceg R, Brown KK. Nintedanib in progressive fibrosing interstitial lung diseases. N Engl J Med. 2019;381(18):1718–27.
Slaine PD, Kleer M, Smith NK, Khaperskyy DA, McCormick C. Stress granule-inducing eukaryotic translation initiation factor 4A inhibitors block influenza A virus replication. Viruses. 2017;9(12):388.
Fan W, Qian S, Qian P, Li X. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016;220:112–6.
Yan H, Ma L, Wang H, Wu S, Huang H, Gu Z, Jiang J, Li Y. Luteolin decreases the yield of influenza A virus in vitro by interfering with the coat protein I complex expression. J Nat Med. 2019;73(3):487–96.
Yi L, et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol. 2004;78(20):11334–9.
Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020;35(1):145–51.
Khaerunnisa S, Kurniawan H, Awaluddin R, Suhartati S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints. 2020;39(8):2000028.
Ton AT, Gentile F, Hsing M, Ban F, Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol Inform. 2020;39(8):2000028.
Smith M. Repurposing therapeutics for COVID-19: supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface. Chem-Rxiv 2020.
Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect. 2020;53(3):368–70.
Stebbing J, Krishnan V, de Bono S, et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol Med. 2020;12:e12697. https://doi.org/10.15252/emmm.202012697.
Kim Y, Jedrzejczak R, Maltseva N, Endres M, Godzik A, Michalska K, Joachimiak A. Crystal structure of NSP15 endoribonuclease from SARS CoV-2. Protein Sci. 2020;29(7):1596–605.
Funding
M.S. was financially supported by the Dr Sulaiman Al Habib Research Center under an Excellence Award (Project Reference Number: COVID-19 RC#09).
Author information
Authors and Affiliations
Contributions
ARS, MS, and SI: study conceptualization and writing (review and editing) the manuscript. RA and HA: data curation, formal analysis, and writing (original draft). MS: funding acquisition and project administration. SI and MS: supervision of the project. ARS, RA, HA, LV, and MS: formal analysis and writing (original draft).
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest.
Ethical approval
No ethical issues exist in the article, and all authors approve publication.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Afreen, R., Iqbal, S., Shah, A.R. et al. In Silico Identification of Potential Inhibitors of the SARS-CoV-2 Nucleocapsid Through Molecular Docking-Based Drug Repurposing. Dr. Sulaiman Al Habib Med J 4, 64–76 (2022). https://doi.org/10.1007/s44229-022-00004-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s44229-022-00004-z