Abstract
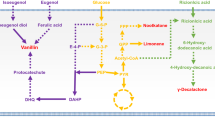
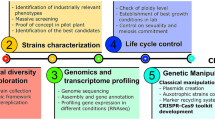
Rudimentary food fermentation can be defined as a spontaneous process of conversion of food components through enzymatic action. A great variety of fermented foods are produced using spontaneous approaches; however, cocoa and coffee represent the most important agricultural commodities on international markets. As a manner to increase the efficiency of these processes, starter cultures have been developed and applied under field conditions. The selection process begins with the recovery of microbial strains from spontaneous fermentation through phenotypic and metabolic traits. Next, mutation-based breeding is used to develop and improve well-adapted starter cultures. With advances in synthetic biology, especially in the last decade, the development of robust cellular fabrications with high fermentative capacity has become easier—largely due to the development of genomic approaches, such as next-generation sequencing techniques, CRISPR-Cas system and bioinformatics tools. This review brings prospects on the use of synthetic biology to design new robust strains for use in cocoa and coffee fermentations, but which can be extended to other rudimentary foods. In addition, metabolic traits and target genes (e.g., UvrA, RecA, GPD1, and GPP2) are proposed as a starting point for the improvement of cocoa and coffee starters. Finally, the regulatory and safety requirements for these food crops are addressed. This review aims to stimulate research on the process of fermentation and the associated synthetic biology tools to produce fermented food efficiently and sustainably.

Similar content being viewed by others
References
Liu L, Wang J, Levin MJ, Sinnott-Armstrong N, Zhao H, Zhao Y, Shao J, Di N, Zhang T. The origins of specialized pottery and diverse alcohol fermentation techniques in early neolithic China. Proc Natl Acad Sci U S A. 2019;116:12767–74. https://doi.org/10.1073/pnas.1902668116.
Tamang JP, Cotter PD, Endo A, Han NS, Kort R, Liu SQ, Mayo B, Westerik N, Hutkins R. Fermented foods in a global age: east meets west. Compr Rev Food Sci Food Saf. 2020;19:184–217. https://doi.org/10.1111/1541-4337.12520.
Shoda S. Seeking prehistoric fermented food in Japan and Korea. Curr Anthropol. 2021;62:S242–55. https://doi.org/10.1086/715808.
Craig OE. Prehistoric fermentation, delayed-return economies, and the adoption of pottery technology. Curr Anthropol. 2021;62:S233–41. https://doi.org/10.1086/716610.
Soni S, Dey G. Perspectives on global fermented foods. Br Food J. 2014;116:1767–87. https://doi.org/10.1108/BFJ-01-2014-0032.
Anal AK. Quality ingredients and safety concerns for traditional fermented foods and beverages from Asia: a review. Fermentation. 2019;5:8. https://doi.org/10.3390/fermentation5010008.
de Pereira GVM, de Carvalho Neto DP, Maske BL, de Dea Lindner J, Vale AS, Favero GR, Viesser J, de Carvalho JC, Góes-Neto A, Soccol CR. An updated review on bacterial community composition of traditional fermented milk products: what next-generation sequencing has revealed so far? Crit Rev Food Sci Nutr. 2022;62:1870–89.
Vale AS, Pereira GVM, Carvalho-Neto DP, Sorto RD, Goés-Neto A, Kato R, Soccol CR. Facility-specific ‘house’ microbiome ensures the maintenance of functional microbial communities into coffee beans fermentation: implications for source tracking. Environ Microbiol Rep. 2021;13:470–81. https://doi.org/10.1111/1758-2229.12921.
Pacheco-Montealegre ME, Dávila-Mora LL, Botero-Rute LM, Reyes A, Caro-Quintero A. Fine resolution analysis of microbial communities provides insights into the variability of cocoa bean fermentation. Front Microbiol. 2020;11:650. https://doi.org/10.3389/fmicb.2020.00650.
de Junqueira ACO, de Pereira GVM, Coral-Medina JD, Alvear MCR, Rosero R, de Carvalho-Neto DP, Enríquez HG, Soccol CR. First description of bacterial and fungal communities in Colombian coffee beans fermentation analysed using illumina-based amplicon sequencing. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-45002-8.
Tamang JP, Watanabe K, Holzapfel WH. Review: diversity of microorganisms in global fermented foods and beverages. Front Microbiol. 2016. https://doi.org/10.3389/fmicb.2016.00377.
Pereira GVDM, de Carvalho-Neto DP, Junqueira ACDO, Karp SG, Letti LAJ, Magalhães-Júnior AI, Soccol CR. A review of selection criteria for starter culture development in the food fermentation industry. Food Rev Int. 2020;36:135–67.
Cardinale S, Arkin AP. Contextualizing context for synthetic biology—identifying causes of failure of synthetic biological systems. Biotechnol J. 2012;7:856–66.
Chen B, Lee HL, Heng YC, Chua N, Teo WS, Choi WJ, Leong SSJ, Foo JL, Chang MW. Synthetic biology toolkits and applications in saccharomyces cerevisiae. Biotechnol Adv. 2018;36:1870–81.
Kunjapur AM, Pfingstag P, Thompson NC. Gene synthesis allows biologists to source genes from farther away in the tree of life. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-06798-7.
Tan X, Letendre JH, Collins JJ, Wong WW. Synthetic biology in the clinic: engineering vaccines, diagnostics, and therapeutics. Cell. 2021;184:881–98.
Vavitsas K, Kugler A, Satta A, Hatzinikolaou DG, Lindblad P, Fewer DP, Lindberg P, Toivari M, Stensjö K. Doing synthetic biology with photosynthetic microorganisms. Physiol Plant. 2021;173:624–38. https://doi.org/10.1111/ppl.13455.
Lv X, Wu Y, Gong M, Deng J, Gu Y, Liu Y, Li J, Du G, Ledesma-Amaro R, Liu L, et al. Synthetic biology for future food: research progress and future directions. Fut Foods. 2021;3:100025.
Predan GMI, Lazăr DA, Lungu II. Cocoa industry-from plant cultivation to cocoa drinks production. In: Grumezescu AM, Holban AM, editors. Caffeinated and cocoa based beverages. Boca Raton: Academic Press; 2019. p. 489–507 (ISBN 9780128158647).
International Cocoa Organization (ICCO) Data on Production and Grindings of Cocoa Beans Available online: https://www.icco.org/ accessed on 6 Jun 2022.
Vásquez ZS, Carvalho DP, Pereira GVM, Vandenberghe LPS, de Oliveira PZ, Tiburcio PB, Rogez HLG, Góes A, Soccol CR. Biotechnological approaches for cocoa waste management: a review. Waste Manag. 2019;90:72–83. https://doi.org/10.1016/j.wasman.2019.04.030.
Viesser JA, de Melo-Pereira GV, de Carvalho-Neto DP, Favero GR, de Carvalho JC, Goés-Neto A, Rogez H, Soccol CR. Global cocoa fermentation microbiome: revealing new taxa and microbial functions by next generation sequencing technologies. World J Microbiol Biotechnol. 2021. https://doi.org/10.1007/s11274-021-03079-2.
Afoakwa EO. Chocolate science and technology. Chinchester: Wiley-Blackwell; 2010. (ISBN 9781405199063).
Fowler MS. Cocoa beans: from tree to factory. 4th ed. Chinchester: Blackwell Publishing; 2009. (ISBN 1405139498 Beckett, S.T., Ed).
Papalexandratou Z, Kaasik K, Kauffmann LV, Skorstengaard A, Bouillon G, Espensen JL, Hansen LH, Jakobsen RR, Blennow A, Krych L, et al. Linking cocoa varietals and microbial diversity of nicaraguan fine cocoa bean fermentations and their impact on final cocoa quality appreciation. Int J Food Microbiol. 2019;304:106–18. https://doi.org/10.1016/j.ijfoodmicro.2019.05.012.
Figueroa-Hernández C, Mota-Gutierrez J, Ferrocino I, Hernández-Estrada ZJ, González-Ríos O, Cocolin L, Suárez-Quiroz ML. The challenges and perspectives of the selection of starter cultures for fermented cocoa beans. Int J Food Microbiol. 2019;301:41–50.
De Vuyst L, Weckx S. The cocoa bean fermentation process: from ecosystem analysis to starter culture development. J Appl Microbiol. 2016;121:5–17. https://doi.org/10.1111/jam.13045.
De Vuyst L, Leroy F. Functional role of yeasts, lactic acid bacteria and acetic acid bacteria in cocoa fermentation processes. FEMS Microbiol Rev. 2020;44:432–53. https://doi.org/10.1093/femsre/fuaa014.
Schwan RF, Wheals AE. The microbiology of cocoa fermentation and its role in chocolate quality. Crit Rev Food Sci Nutr. 2004;44:205–21. https://doi.org/10.1080/10408690490464104.
Illeghems K, de Vuyst L, Papalexandratou Z, Weckx S. Phylogenetic analysis of a spontaneous cocoa bean fermentation metagenome reveals new insights into its bacterial and fungal community diversity. PLoS ONE. 2012;7:e38040. https://doi.org/10.1371/journal.pone.0038040.
Hamdouche Y, Meile JC, Lebrun M, Guehi T, Boulanger R, Teyssier C, Montet D. Impact of turning, pod storage and fermentation time on microbial ecology and volatile composition of cocoa beans. Food Res Int. 2019;119:477–91. https://doi.org/10.1016/j.foodres.2019.01.001.
Pereira GVM, Magalhães-Guedes KT, Schwan RF. RDNA-based DGGE analysis and electron microscopic observation of cocoa beans to monitor microbial diversity and distribution during the fermentation process. Food Res Int. 2013;53:482–6. https://doi.org/10.1016/j.foodres.2013.05.030.
Jespersen L, Nielsen D, Honholt S, Jakobsen M. Occurrence and diversity of yeasts involved in fermentation of west African cocoa beans. FEMS Yeast Res. 2005;5:441–53. https://doi.org/10.1016/j.femsyr.2004.11.002.
Mota-Gutierrez J, Botta C, Ferrocino I, Giordano M, Bertolino M, Dolci P, Cannoni M, Cocolin L. Dynamics and biodiversity of bacterial and yeast communities during fermentation of cocoa beans. Appl Environ Microbiol. 2018;84:e01164-e1218. https://doi.org/10.1128/AEM.01164-18.
Daniel HM, Vrancken G, Takrama JF, Camu N, De Vos P, De Vuyst L. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 2009;9:774–83. https://doi.org/10.1111/j.1567-1364.2009.00520.x.
Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. The microbiology of ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol. 2007;114:168–86. https://doi.org/10.1016/j.ijfoodmicro.2006.09.010.
Serra JL, Moura FG, de Pereira GVM, Soccol CR, Rogez H, Darnet S. Determination of the microbial community in Amazonian cocoa bean fermentation by illumina-based metagenomic sequencing. Lwt. 2019;106:229–39. https://doi.org/10.1016/j.lwt.2019.02.038.
Hernández-Hernández C, López-Andrade PA, Ramírez-Guillermo MA, Guerra Ramírez D, Caballero Pérez JF. Evaluation of different fermentation processes for use by small cocoa growers in Mexico. Food Sci Nutr. 2016;4:690–5. https://doi.org/10.1002/fsn3.333.
Lefeber T, Papalexandratou Z, Gobert W, Camu N, De Vuyst L. On-farm implementation of a starter culture for improved cocoa bean fermentation and its influence on the flavour of chocolates produced thereof. Food Microbiol. 2012;30:379–92. https://doi.org/10.1016/j.fm.2011.12.021.
Hamdouche Y, Guehi T, Durand N, Kedjebo KBD, Montet D, Meile JC. Dynamics of microbial ecology during cocoa fermentation and drying: towards the identification of molecular markers. Food Control. 2015. https://doi.org/10.1016/j.foodcont.2014.05.031.
Viesser JA, de Melo-Pereira GV, de Carvalho-Neto DP, Favero GR, de Carvalho JC, Goés-Neto A, Rogez H, Soccol CR. Global cocoa fermentation microbiome: revealing new taxa and microbial functions by next generation sequencing technologies. World J Microbiol Biotechnol. 2021;37:118. https://doi.org/10.1007/s11274-021-03079-2.
Adler P, Bolten CJ, Dohnt K, Hansen CE, Wittmann C. Core fluxome and metafluxome of lactic acid bacteria under simulated cocoa pulp fermentation conditions. Appl Environ Microbiol. 2013;79:5670–81. https://doi.org/10.1128/AEM.01483-13.
Camu N, De Winter T, Verbrugghe K, Cleenwerck I, Vandamme P, Takrama JS, Vancanneyt M, De Vuyst L. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl Environ Microbiol. 2007;73:1809–24. https://doi.org/10.1128/AEM.02189-06.
Ho VTT, Zhao J, Fleet G. The effect of lactic acid bacteria on cocoa bean fermentation. Int J Food Microbiol. 2015;205:54–67. https://doi.org/10.1016/j.ijfoodmicro.2015.03.031.
Oestreich-Janzen S. Chemistry of coffee. Elsevier Inc.; 2013. (ISBN 9780124095472).
de Pereira GVM, Soccol VT, Soccol CR. Current state of research on cocoa and coffee fermentations. Curr Opin Food Sci. 2016;7:50–7.
Huch M, Franz CMAP. Coffee: fermentation and microbiota. Woodhead Publishing; 2015. (ISBN 9781782420156).
Elhalis H, Cox J, Zhao J. Ecological diversity, evolution and metabolism of microbial communities in the wet fermentation of Australian coffee beans. Int J Food Microbiol. 2020. https://doi.org/10.1016/j.ijfoodmicro.2020.108544.
Pereira TS, Batista NN, Santos-Pimenta LP, Martinez SJ, Ribeiro LS, Oliveira-Naves JA, Schwan RF. Self-induced anaerobiosis coffee fermentation: impact on microbial communities, chemical composition and sensory quality of coffee. Food Microbiol. 2022. https://doi.org/10.1016/j.fm.2021.103962.
Pothakos V, De Vuyst L, Zhang SJ, De Bruyn F, Verce M, Torres J, Callanan M, Moccand C, Weckx S. Temporal shotgun metagenomics of an Ecuadorian coffee fermentation process highlights the predominance of lactic acid bacteria. Curr Res Biotechnol. 2020;2:1–15. https://doi.org/10.1016/j.crbiot.2020.02.001.
Lee LW, Cheong MW, Curran P, Yu B, Liu SQ. Coffee fermentation and flavour—an intricate and delicate relationship. Food Chem. 2015;185:182–91. https://doi.org/10.1016/j.foodchem.2015.03.124.
Evangelista SR, da Miguel MGCP, Silva CF, Pinheiro ACM, Schwan RF. Microbiological diversity associated with the spontaneous wet method of coffee fermentation. Int J Food Microbiol. 2015;210:102–12. https://doi.org/10.1016/j.ijfoodmicro.2015.06.008.
Avallone S, Brillouet JM, Guyot B, Olguin E, Guiraud JP. Involvement of pectolytic micro-organisms in coffee fermentation. Int J Food Sci Technol. 2002;37:191–8. https://doi.org/10.1046/j.1365-2621.2002.00556.x.
Djossou O, Perraud-Gaime I, Mirleau FL, Rodriguez-Serrano G, Karou G, Niamke S, Ouzari I, Boudabous A, Roussos S. Robusta coffee beans post-harvest microflora: Lactobacillus plantarum sp. as potential antagonist of Aspergillus carbonarius. Anaerobe. 2011;17:267–72. https://doi.org/10.1016/j.anaerobe.2011.03.006.
de Pereira GVM, de Carvalho-Neto DP, Medeiros ABP, Soccol VT, Neto E, Woiciechowski AL, Soccol CR. Potential of lactic acid bacteria to improve the fermentation and quality of coffee during on-farm processing. Int J Food Sci Technol. 2016;51:1689–95. https://doi.org/10.1111/ijfs.13142.
de Carvalho-Neto DP, de Melo-Pereira GV, Finco AMO, Letti LAJ, da Silva BJG, Vandenberghe LPS, Soccol CR. Efficient coffee beans mucilage layer removal using lactic acid fermentation in a stirred-tank bioreactor: kinetic metabolic and sensorial studies. Food Biosci. 2018;26:80–7. https://doi.org/10.1016/j.fbio.2018.10.005.
de Pereira GVM, da Silva-Vale A, de Carvalho-Neto DP, Muynarsk ES, Soccol VT, Soccol CR. Lactic acid bacteria: what coffee industry should know? Curr Opin Food Sci. 2020;31:1–8.
De Bruyn F, Zhang SJ, Pothakos V, Torres J, Lambot C, Moroni AV, Callanan M, Sybesma W, Weckx S, Vuyst LD. Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production. Appl Environ Microbiol. 2017;83:e02398-e2416 (0099-2240).
de Pereira GVM, de Carvalho-Neto DP, Magalhães-Júnior AI, Vásquez ZS, Medeiros ABP, Vandenberghe LPS, Soccol CR. Exploring the impacts of postharvest processing on the aroma formation of coffee beans—a review. Food Chem. 2019;272:441–52.
Hazelwood LA, Daran JM, Van Maris AJA, Pronk JT, Dickinson JR. The ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol. 2008;74:2259–66. https://doi.org/10.1128/AEM.02625-07.
Bachmann H, Pronk JT, Kleerebezem M, Teusink B. Evolutionary engineering to enhance starter culture performance in food fermentations. Curr Opin Biotechnol. 2015;32:1–7.
Bizaj E, Cordente AG, Bellon JR, Raspor P, Curtin CD, Pretorius IS. A breeding strategy to harness flavor diversity of saccharomyces interspecific hybrids and minimize hydrogen sulfide production. FEMS Yeast Res. 2012;12:456–65. https://doi.org/10.1111/j.1567-1364.2012.00797.x.
Steensels J, Snoek T, Meersman E, Nicolino MP, Voordeckers K, Verstrepen KJ. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol Rev. 2014;38:947–95. https://doi.org/10.1111/1574-6976.12073.
Schwan RF, Cooper RM, Wheals AE. Endopolygalacturonase secretion by Kluyveromyces marxianus and other cocoa pulp-degrading yeasts. Enzyme Microb Technol. 1997;21:234.
Leal GA, Gomes LH, Efraim P, de Almeida Tavares FC, Figueira A. Fermentation of cacao (Theobroma cacao L) Seeds with a hybrid Kluyveromyces marxianus strain improved product quality attributes. FEMS Yeast Res. 2008;8:788–98. https://doi.org/10.1111/j.1567-1364.2008.00405.x.
Meersman E, Steensels J, Paulus T, Struyf N, Saels V, Mathawan M, Koffi J, Vrancken G, Verstrepena KJ. Breeding strategy to generate robust yeast starter cultures for cocoa pulp fermentations. Appl Environ Microbiol. 2015;81:6166–76. https://doi.org/10.1128/AEM.00133-15.
Choi KR, Jang WD, Yang D, Cho JS, Park D, Lee SY. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019;37:817–37.
Liu L, Guan N, Li J, Shin HD, Du G, Chen J. Development of GRAS strains for nutraceutical production using systems and synthetic biology approaches: advances and prospects. Crit Rev Biotechnol. 2017;37:139–50.
Markham KA, Alper HS. Synthetic biology expands the industrial potential of yarrowia lipolytica. Trends Biotechnol. 2018;36:1085–95.
Opgenorth P, Costello Z, Okada T, Goyal G, Chen Y, Gin J, Benites V, de Raad M, Northen TR, Deng K, et al. Lessons from two design-build-test-learn cycles of dodecanol production in Escherichia coli aided by machine learning. ACS Synth Biol. 2019;8:1337–51. https://doi.org/10.1021/acssynbio.9b00020.
Kong LH, Xiong ZQ, Song X, Xia YJ, Zhang N, Ai LZ. Characterization of a panel of strong constitutive promoters from Streptococcus thermophilus for fine-tuning gene expression. ACS Synth Biol. 2019;8:1469–72. https://doi.org/10.1021/acssynbio.9b00045.
Son J, Jeong KJ. Recent advances in synthetic biology for the engineering of lactic acid bacteria. Biotechnol Bioprocess Eng. 2020;25:962–73.
Monteiro LMO, Arruda LM, Silva-Rocha R. Emergent properties in complex synthetic bacterial promoters. ACS Synth Biol. 2018;7:602–12. https://doi.org/10.1021/acssynbio.7b00344.
Kiesenhofer DP, Mach RL, Mach-Aigner AR. Influence of cis element arrangement on promoter strength in Trichoderma reesei. Appl Environ Microbiol. 2018. https://doi.org/10.1128/AEM.
Westmann CA, de Alves LF, Silva-Rocha R, Guazzaroni ME. Mining novel constitutive promoter elements in soil metagenomic libraries in Escherichia coli. Front Microbiol. 2018. https://doi.org/10.3389/fmicb.2018.01344.
Lee D, Lloyd NDR, Pretorius IS, Borneman AR. Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion. Microb Cell Factories. 2016. https://doi.org/10.1186/s12934-016-0446-2.
Salem FH, Lebrun M, Mestres C, Sieczkowski N, Boulanger R, Collignan A. Transfer kinetics of labeled aroma compounds from liquid media into coffee beans during simulated wet processing conditions. Food Chem. 2020. https://doi.org/10.1016/j.foodchem.2020.126779.
Barrangou R, Marraffini LA. CRISPR-cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–44.
Song X, Zhang X, Xiong Y, Qiang Z, Liu X, Xia YJ, Wang SJ, Ai LZ. CRISPR–cas-mediated gene editing in lactic acid bacteria. Mol Biol Rep. 2020;47:8133–44.
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264.
Vicente MM, Chaves-Ferreira M, Jorge JMP, Proença JT, Barreto VM. The off-targets of clustered regularly interspaced short palindromic repeats gene editing. Front Cell Dev Biol. 2021;9:718466.
Shen W, Zhang J, Geng B, Qiu M, Hu M, Yang Q, Bao W, Xiao Y, Zheng Y, Peng W, et al. Establishment and application of a CRISPR-cas12a assisted genome-editing system in Zymomonas mobilis. Microb Cell Factories. 2019. https://doi.org/10.1186/s12934-019-1219-5.
Muysson J, Miller L, Allie R, Inglis DL. The use of CRISPR-Cas9 genome editing to determine the importance of glycerol uptake in wine yeast during icewine fermentation. Fermentation. 2019. https://doi.org/10.3390/fermentation5040093.
Jensen ED, Ferreira R, Jakočiunas T, Arsovska D, Zhang J, Ding L, Smith JD, David F, Nielsen J, Jensen MK, et al. Transcriptional reprogramming in yeast using DCas9 and combinatorial GRNA strategies. Microb Cell Factories. 2017. https://doi.org/10.1186/s12934-017-0664-2.
Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer PRE, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–8. https://doi.org/10.1038/nmeth.3312.
Muynarsk ESM, de Melo-Pereira GV, Mesa D, Thomaz-Soccol V, Carvalho JC, Pagnoncelli MGB, Soccol CR. Draft genome sequence of Pediococcus acidilactici strain LPBC161, isolated from mature coffee cherries during natural fermentation. Microbiol Resour Announc. 2019. https://doi.org/10.1128/mra.00332-19.
Illeghems K, de Vuyst L, Weckx S. Comparative genome analysis of the candidate functional starter culture strains Lactobacillus fermentum 222 and Lactobacillus plantarum 80 for controlled cocoa bean fermentation processes. BMC Genomics. 2015. https://doi.org/10.1186/s12864-015-1927-0.
Illeghems K, de Vuyst L, Weckx S. Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics. 2013. https://doi.org/10.1186/1471-2164-14-526.
Bauer FF, Pretorius IS. Yeast stress response and fermentation efficiency: how to survive the making of wine—a review. SAJEV. 2000. https://doi.org/10.21548/21-1-3557.
Elhalis H, Cox J, Frank D, Zhao J. Microbiological and biochemical performances of six yeast species as potential starter cultures for wet fermentation of coffee beans. LWT. 2021. https://doi.org/10.1016/j.lwt.2020.110430.
de Pereira GVM, da Miguel MGCP, Ramos CL, Schwan RF. Microbiological and physicochemical characterization of small-scale cocoa fermentations and screening of yeast and bacterial strains to develop a defined starter culture. Appl Environ Microbiol. 2012;78:5395–405. https://doi.org/10.1128/AEM.01144-12.
Visintin S, Alessandria V, Valente A, Dolci P, Cocolin L. Molecular identification and physiological characterization of yeasts, lactic acid bacteria and acetic acid bacteria isolated from heap and box cocoa bean fermentations in West Africa. Int J Food Microbiol. 2016;216:69–78. https://doi.org/10.1016/j.ijfoodmicro.2015.09.004.
Díaz-Muñoz C, de Vuyst L. Functional yeast starter cultures for cocoa fermentation. J Appl Microbiol. 2021;133:39–66.
Gonçalves M, Pontes A, Almeida P, Barbosa R, Serra M, Libkind D, Hutzler M, Gonçalves P, Sampaio JP. Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr Biol. 2016;26:2750–61. https://doi.org/10.1016/j.cub.2016.08.040.
Kostopoulou A, Batrinou A. Of gene expression of saccharomyces cerevisiae under osmotic stress in fermentation processes; Article, 2018; Vol. 13.
de Pereira GVM, Neto E, Soccol VT, Medeiros ABP, Woiciechowski AL, Soccol CR. Conducting starter culture-controlled fermentations of coffee beans during on-farm wet processing: growth, metabolic analyses and sensorial effects. Food Res Int. 2015;75:348–56. https://doi.org/10.1016/j.foodres.2015.06.027.
van de Guchte M, Serror P, Chervaux C, Smokvina T, Ehrlich SD, Maguin E. Stress responses in lactic acid bacteria, 82; 2002.
Grinholc M, Rodziewicz A, Forys K, Rapacka-Zdonczyk A, Kawiak A, Domachowska A, Golunski G, Wolz C, Mesak L, Becker K, et al. Fine-tuning RecA expression in Staphylococcus aureus for antimicrobial photoinactivation: importance of photo-induced DNA damage in the photoinactivation mechanism. Appl Microbiol Biotechnol. 2015;99:9161–76. https://doi.org/10.1007/s00253-015-6863-z.
Zheng Y, Wang J, Bai X, Chang Y, Mou J, Song J, Wang M. Improving the acetic acid tolerance and fermentation of Acetobacter pasteurianus by nucleotide excision repair protein UvrA. Appl Microbiol Biotechnol. 2018;102:6493–502. https://doi.org/10.1007/s00253-018-9066-6.
Samagaci L, Ouattara H, Niamké S, Lemaire M. Pichia kudrazevii and candida nitrativorans are the most well-adapted and relevant yeast species fermenting cocoa in Agneby-Tiassa, a local Ivorian cocoa producing region. Food Res Int. 2016;89:773–80. https://doi.org/10.1016/j.foodres.2016.10.007.
Silva CF, Vilela DM, de Souza Cordeiro C, Duarte WF, Dias DR, Schwan RF. Evaluation of a potential starter culture for enhance quality of coffee fermentation. World J Microbiol Biotechnol. 2013;29:235–47. https://doi.org/10.1007/s11274-012-1175-2.
de Pereira GVM, Soccol VT, Pandey A, Medeiros ABP, Andrade-Lara JMR, Gollo AL, Soccol CR. Isolation, selection and evaluation of yeasts for use in fermentation of coffee beans by the wet process. Int J Food Microbiol. 2014;188:60–6. https://doi.org/10.1016/j.ijfoodmicro.2014.07.008.
Delgado-Ospina J, Triboletti S, Alessandria V, Serio A, Sergi M, Paparella A, Rantsiou K, Chaves-López C. Functional biodiversity of yeasts isolated from Colombian fermented and dry cocoa beans. Microorganisms. 2020;8:1–17. https://doi.org/10.3390/microorganisms8071086.
de Junqueira ACO, de Vinícius-Melo PG, Viesser JA, de Carvalho-Neto DP, Querne LBP, Soccol CR. Isolation and selection of fructose-consuming lactic acid bacteria associated with coffee bean fermentation. Food Biotechnol. 2022;36:58–75. https://doi.org/10.1080/08905436.2021.2007119.
Ouattara HG, Elias RJ, Dudley EG. Microbial synergy between Pichia kudriazevii YS201 and Bacillus subtilis BS38 improves pulp degradation and aroma production in cocoa pulp simulation medium. Heliyon. 2020. https://doi.org/10.1016/j.heliyon.2020.e03269.
Lee AH, Neilson AP, O’Keefe SF, Ogejo JA, Huang H, Ponder M, Chu HSS, Jin Q, Pilot G, Stewart AC. A laboratory-scale model cocoa fermentation using dried, unfermented beans and artificial pulp can simulate the microbial and chemical changes of on-farm cocoa fermentation. Eur Food Res Technol. 2019;245:511–9. https://doi.org/10.1007/s00217-018-3171-8.
Lefeber T, Janssens M, Camu N, de Vuyst L. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl Environ Microbiol. 2010;76:7708–16. https://doi.org/10.1128/AEM.01206-10.
Zeng X, Jiang H, Yang G, Ou Y, Lu S, Jiang J, Lei R, Su L. Regulation and management of the biosecurity for synthetic biology. Syn Syst Biotechnol. 2022;7:784–90. https://doi.org/10.1016/j.synbio.2022.03.005.
Laulund S, Wind A, Derkx P, Zuliani V. Regulatory and safety requirements for food cultures. Microorganisms. 2017;5:28. https://doi.org/10.3390/microorganisms5020028.
Laranjo M, Elias M, Fraqueza MJ. The use of starter cultures in traditional meat products. J Food Qual. 2017;2017:1–18. https://doi.org/10.1155/2017/9546026.
Teng TS, Chin YL, Chai KF, Chen WN. Fermentation for future food systems. EMBO Rep. 2021. https://doi.org/10.15252/embr.202152680.
Li J, Zhao H, Zheng L, An W. Advances in synthetic biology and biosafety governance. Front Bioeng Biotechnol. 2021. https://doi.org/10.3389/fbioe.2021.598087.
Sajid M, Stone SR, Kaur P. Transforming traditional nutrition paradigms with synthetic biology driven microbial production platforms. Curr Res Biotechnol. 2021;3:260–8. https://doi.org/10.1016/j.crbiot.2021.07.002.
Jagtap UB, Jadhav JP, Bapat VA, Pretorius IS. Synthetic biology stretching the realms of possibility in wine yeast research. Int J Food Microbiol. 2017;252:24–34. https://doi.org/10.1016/j.ijfoodmicro.2017.04.006.
Alperstein L, Gardner JM, Sundstrom JF, Sumby KM, Jiranek V. Yeast Bioprospecting versus synthetic biology—which is better for innovative beverage fermentation? Appl Microbiol Biotechnol. 2020;104:1939–53. https://doi.org/10.1007/s00253-020-10364-x.
Lallemand Sourvisiae Technical Data Sheet Available online: https://www.lallemandbrewing.com/wp-content/uploads/2019/09/LAL-TDS-Mascoma-Sourvisiae-Final-1.pdf accessed on 4 Jun 2022
Morrissey JP, Etschmann MMW, Schrader J, de Billerbeck GM. Cell factory applications of the yeast Kluyveromyces Marxianus for the biotechnological production of natural flavour and fragrance molecules. Yeast. 2015;32:3–16. https://doi.org/10.1002/yea.3054.
van Wyk N, Kroukamp H, Pretorius I. The smell of synthetic biology: engineering strategies for aroma compound production in yeast. Fermentation. 2018;4:54. https://doi.org/10.3390/fermentation4030054.
Shukla P. Synthetic biology perspectives of microbial enzymes and their innovative applications. Indian J Microbiol. 2019;59:401–9. https://doi.org/10.1007/s12088-019-00819-9.
Randall A, Guye P, Gupta S, Duportet X, Weiss R. Design and connection of robust genetic circuits. Elsevier; 2011. p. 159–86.
Davenport BI, Tica J, Isalan M. Reducing metabolic burden in the PACEmid evolver system by remastering high-copy phagemid vectors. Eng Biol. 2022. https://doi.org/10.1049/enb2.12021.
Kumar V, Hart AJ, Keerthiraju ER, Waldron PR, Tucker GA, Greetham D. Expression of mitochondrial cytochrome c oxidase chaperone gene (COX20) improves tolerance to weak acid and oxidative stress during yeast fermentation. PLoS ONE. 2015. https://doi.org/10.1371/journal.pone.0139129.
Tian H, Tan J, Zhang L, Gu X, Xu W, Guo X, Luo Y. Increase of stress resistance in Lactococcus lactis via a novel food-grade vector expressing a shsp gene from Streptococcus thermophilus. Braz J Microbiol. 2012;43:1157–64.
Dank A, Smid EJ, Notebaart RA. CRISPR-cas genome engineering of esterase activity in Saccharomyces cerevisiae steers aroma formation. BMC Res Notes. 2018. https://doi.org/10.1186/s13104-018-3788-5.
Varela C, Kutyna DR, Solomon MR, Black CA, Borneman A, Henschke PA, Pretorius IS, Chambers PJ. Evaluation of gene modification strategies for the development of low-alcohol-wine yeasts. Appl Environ Microbiol. 2012;78:6068–77. https://doi.org/10.1128/AEM.01279-12.
Pardo E, Rico J, Gil JV, Orejas M. De novo production of six key grape aroma monoterpenes by a geraniol synthase-engineered S. cerevisiae wine strain. Microb Cell Factories. 2015. https://doi.org/10.1186/s12934-015-0306-5.
Mertens S, Gallone B, Steensels J, Herrera-Malaver B, Cortebeek J, Nolmans R, Saels V, Vyas VK, Verstrepen KJ. Reducing phenolic off-flavors through CRISPR-based gene editing of the FDC1 gene in Saccharomyces cerevisiae x Saccharomyces eubayanus hybrid lager beer yeasts. PLoS ONE. 2019. https://doi.org/10.1371/journal.pone.0209124.
van Wyk N, Kroukamp H, Espinosa MI, von Wallbrunn C, Wendland J, Pretorius IS. Blending wine yeast phenotypes with the aid of CRISPR DNA editing technologies. Int J Food Microbiol. 2020. https://doi.org/10.1016/j.ijfoodmicro.2020.108615.
Funding
This study is funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico, 306421/2020-8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
da Silva Vale, A., de Melo Pereira, G.V., Santana, L.M. et al. Perspective on the use of synthetic biology in rudimentary food fermentations. Syst Microbiol and Biomanuf 3, 150–165 (2023). https://doi.org/10.1007/s43393-022-00131-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00131-6