Abstract
Papillomaviruses (PVs) have been identified in several animal species, including dogs (canine papillomaviruses, CPVs) and cattle (bovine papillomaviruses, BPVs). Although some BPVs may occasionally infect species other than cattle, to the best of our knowledge, BPVs have not been reported in dogs to date. Herein, we carried out a retrospective phylogenetic study of PVs circulating in dogs from southern Brazil between 2017 and 2022, also investigating possible mixed infections and spillover events. For this, we screened 32 canine papilloma samples by PCR using the degenerate primers FAP59/64 and/or MY09/11, which amplify different regions of the L1 gene; the genomic target often used for PV classification/typing. Out these, 23 PV DNA samples were successfully amplified and sequenced. All PVs amplified by FAP59/64 (n = 22) were classified as CPV-1. On the other hand, PVs amplified by MY09/11 (n = 4) were classified as putative BPV-1. Among these, three samples showed mixed infection by CPV-1 and putative BPV-1. One of the putative BPV-1 detected in co-infected samples had the L1 gene full-sequenced, confirming the gene identity. Furthermore, the phylogenetic classifications from the FAP59/64 and/or MY09/11 amplicons were supported by a careful in silico analysis, which demonstrated that the analysis based on them matches to the classification from the complete L1 gene. Overall, we described CPV-1 circulation in southern Brazil over the years and the potencial BPV infection in dogs (potential spillover event), as well as possible CPV/1/BPV-1 co-infections. Finally, we suggest the analysis of the complete genome of the putative BPVs detected in dogs in order to deepen the knowledge about the PV-host interactions.



Similar content being viewed by others
Data availability
The data that support the study findings may be available upon reasonable request to the corresponding author.
References
Rector A, Van Ranst M (2013) Animal Papillomaviruses. Virology 445:213–223
Bocaneti F, Altamura G, Corteggio A, Velescu E, Roperto F, Borzacchiello G (2016) Bovine Papillomavirus: New Insights into an Old Disease. Transbound Emerg Dis 63:14–23
Bam J, Kumar P, Leishangthem GD, Saikia A, Somvanshi R (2013) Spontaneous Cutaneous Papillomatosis in Yaks and Detection and Quantification of Bovine Papillomavirus-1 and -2. Transbound Emerg Dis 60:475–480
Cutarelli A, De Falco F, Uleri V, Buonavoglia C, Roperto S (2021) The Diagnostic Value of the Droplet Digital PCR for the Detection of Bovine Deltapapillomavirus in Goats by Liquid Biopsy. Transbound Emerg Dis 68:3624–3630
Munday JS, Thomson N, Dunowska M, Knight CG, Laurie RE, Hills S (2015) Genomic Characterisation of the Feline Sarcoid-Associated Papillomavirus and Proposed Classification as Bos Taurus Papillomavirus Type 14. Vet Microbiol 177:289–295
Pangty K, Singh S, Goswami R, Saikumar G, Somvanshi R (2010) Detection of BPV-1 and -2 and Quantification of BPV-1 by Real-Time PCR in Cutaneous Warts in Cattle and Buffaloes. Transbound Emerg Dis 57:185–196
Roperto S, Russo V, Corrado F, Munday JS, De Falco F, Roperto F (2018) Detection of Bovine Deltapapillomavirus DNA in Peripheral Blood of Healthy Sheep ( Ovis Aries ). Transbound Emerg Dis 65:758–764
Gil da Costa RM, Peleteiro MC, Pires MA, DiMaio D (2017) An Update on Canine, Feline and Bovine Papillomaviruses Transbound. Emerg Dis 64:1371–1379
Lane HE, Weese JS, Stull JW (2017) Canine Oral Papillomavirus Outbreak at a Dog Daycare Facility. Can Vet J 58:747–749
ICTV Papillomaviridae Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/dsdna-viruses/w/papillomaviridae. Accessed 6 Ago 2023
Papillomavirus Virus Episteme Reference Genomes for Animal Papillomavirus Available online: https://pave.niaid.nih.gov/#explore/reference_genomes/animal_genomes. Accessed 6 Ago 2023
Doorbar J (2005) The Papillomavirus Life Cycle. J Clin Virol 32:7–15
Bernard HU, Burk RD, Chen Z, Van Doorslaer K, ZurHausen H, De Villiers EM (2010) Classification of Papillomaviruses (PVs) Based on 189 PV Types and Proposal of Taxonomic Amendments. Virology 401:70–79
de Villiers E-M, Fauquet C, Broker TR, Bernard H-U (2004) zur Hausen, H. Classification of Papillomaviruses. Virology 324:17–27
PaVE Papillomavirus Virus Episteme: Reference Genomes for Animal Papillomavirus Available online: https://pave.niaid.nih.gov/explore/reference_genomes/animal_genomes Accessed 20 Jul 2023.
Munday JS, Gedye K, Knox MA, Ravens P, Lin X (2022) Genomic Characterisation of Canis Familiaris Papillomavirus Type 24, a Novel Papillomavirus Associated with Extensive Pigmented Plaque Formation in a Pug Dog. Viruses 14:2357
Lange CE, Jennings SH, Diallo A, Lyons J (2019) Canine Papillomavirus Types 1 and 2 in Classical Papillomas: High Abundance, Different Morphological Associations and Frequent Co-Infections. Vet J 250:1–5
Tisza MJ, Yuan H, Schlegel R, Buck CB (2016) Genomic Sequence of Canine Papillomavirus 19. Genome Announc 4:1380–1396. https://doi.org/10.1128/genomeA.01380-16
Forslund O, Antonsson A, Nordin P, Stenquist B, Göran Hansson B (1999) A Broad Range of Human Papillomavirus Types Detected with a General PCR Method Suitable for Analysis of Cutaneous Tumours and Normal Skin. J Gen Virol 80:2437–2443
Manos MM, Ting YT, Wright DK, Lewis AL, Broker TR, Wolinsky SM, Manos MM, Ting YC (1989) Use of Polymerase Chain Reaction Amplification for the Detection of Genital Human Papillomaviruses. Cancer Cells Mol Diagnostics Hum Cancer 7:209–214
Staden R (1996) The Staden Sequence Analysis Package. Mol Biotechnol 5:233–241
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res 22:4673–4680
Hall TA (1999) BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35:1547–1549
Posada D (2008) JModelTest: Phylogenetic Model Averaging. Mol Biol Evol 25:1253–1256
Alcântara BK, Alfieri AA, Rodrigues WB, Otonel RA, Lunardi M, Headley SA, Alfieri AF (2014) Identification of Canine Papillomavirus Type 1 (CPV1) DNA in Dogs with Cutaneous Papillomatosis. Pesqui Veterinária Bras 34(12):1223–1226
Alves CDBT, Weber MN, Guimarães LLB, Cibulski SP, da Silva FRC, Daudt C, Budaszewski RF, Silva MS, Mayer FQ, Bianchi RM et al (2020) Canine Papillomavirus Type 16 Associated to Squamous Cell Carcinoma in a Dog: Virological and Pathological Findings. Brazilian J Microbiol 51:2087–2094
Reis JDR, Oliveira LB, Santos LABO, Soares RC, Batista MVA (2019) Molecular Characterization of Canis Familiaris Oral Papillomavirus 1 Identified in Naturally Infected Dogs from Northeast Brazil. Vet Dermatol 30:424
Merchioratto I, De Oliveira PS, Silva Júnior JV, Brum MC, Weiblen R, Flores EF (2023) Phylogeny and Amino Acid Analysis in Single and Mixed Bovine Papillomavirus Infections in Southern Brazil 2016-2020. Arch Virol 168:52
Silva MAR, Batista MVA, Pontes NE, Santos EUD, Coutinho LCA, Castro RS, Balbino VQ, Freitas AC (2013) Comparison of Two PCR Strategies for the Detection of Bovine Papillomavirus. J Virol Methods 192:55–58
Becker AS, Júnior JV, Weiblen R, Flores EF (2023) An Appraisal of Gene Targets for Phylogenetic Classification of Canine Distemper Virus: Is the Hemagglutinin the Best Candidate? Virus Res. 325:199043
de Oliveira PS, Júnior JV, Weiblen R, Flores EF (2021) Subtyping Bovine Viral Diarrhea Virus (BVDV): Which Viral Gene to Choose? Infect Genet Evol. 92:104891
De Oliveira PS, Silva Júnior JV, Weiblen R, Flores EF (2022) A New (Old) Bovine Viral Diarrhea Virus 2 Subtype: BVDV-2e. Arch Virol 167:2545–2553
Dagalp SB, Dogan F, Farzanı TA, Salar S, Bastan A (2017) The Genetic Diversity of Bovine Papillomaviruses (BPV) from Different Papillomatosis Cases in Dairy Cows in Turkey. Arch Virol 162:1507–1518
Silva MS, Weiss M, Brum MC, Dos Anjos BL, Torres FD, Weiblen R, Flores EF (2010) Molecular Identification of Bovine Papillomaviruses Associated with Cutaneous Warts in Southern Brazil. J Vet Diagnostic Investig 22:603–606
Acknowledgements
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (IM, CIM) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (TRRL, PSBO) funding agencies for scholarships. JVJSJr was supported by Financiadora de Estudos e Projetos (FINEP) DTI-A-1. EFF (process 301414/2010-6) and RW (process 303981/2022-9) were supported by CNPq research fellowships.
Funding
The Programa de Pós-graduação em Medicina Veterinária (UFSM), of which IM, CIM, TRRL and PSBO were/are students, is partially supported by CAPES (Finance code 001).
Author information
Authors and Affiliations
Contributions
IM: conceptualization, experiments, data interpretation, draft and final writing; CIM: experiments and data interpretation; TRRL: experiments and data interpretation; PSBO: experiments and data interpretation; JVJSJr: conceptualization, data interpretation and final writing; MCSB: supply of samples, data interpretation and critical review; RW: data interpretation and critical review; EFF: conceptualization, data interpretation, critical review and supervision.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Consent for publication
All authors approved the version to be published.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Helena Lage Ferreira
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
42770_2024_1349_MOESM2_ESM.pdf
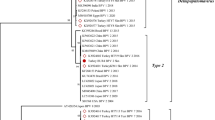
The phylogenetic analysis was performed based on complete L1 sequences (A), complete FAP59/64 (B) and MY09/11 (C), and partial FAP59/64 (D) and MY09/11 (E). Analyses were performed by the Maximum Likelihood method, using the MEGA X software (version 10.2.4) (see Table 1). Bootstrap values were calculated based on 1,000 replicates. The genus Chipapillomavirus is divided into three species (Chi 1, 2 and 3), Lambdapapillomavirus into two species (Lambda 2 and 3) and Taupapillomavirus into two species (Tau 1 and 2), with CPV-21, -22 and -23 not yet have been identified into species. Supplementary file2 (PDF 560 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Merchioratto, I., Mucellini, C.I., Lopes, T.R.R. et al. Phylogenetic analysis of papillomaviruses in dogs from southern Brazil: molecular epidemiology and investigation of mixed infections and spillover events. Braz J Microbiol (2024). https://doi.org/10.1007/s42770-024-01349-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42770-024-01349-3