Abstract
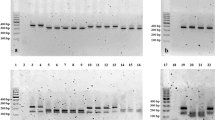
Mycoplasma hyopneumoniae is the etiologic agent of porcine enzootic pneumonia, responsible for major production losses worldwide. The bacteria have a limited metabolism and need to obtain molecules from the growth environment, which causes multiple difficulties for in vitro culture. These limitations have a negative influence on the ability to carry out research for the development of the rational use of antimicrobials and vaccines. The objective of this investigation was to evaluate the genetic profile and in vitro susceptibility of field isolates of M. hyopneumoniae to different antimicrobials. All 16 isolates obtained from the samples presented 100% of identity in the partial sequence of 16S rRNA gene when compared to M. hyopneumoniae. A dendrogram was created using the PCR results of the genes related to pathogenicity, and the isolates were distributed into four clusters, suggesting genetic variability among four different isolates circulating on the same farm. The minimum inhibitory concentration of the isolates was higher for the antimicrobials tylosin (< 0.001–16 mg/L) and spiramycin (< 0.001–16 mg/L) than for enrofloxacin (< 0.001–0.125 mg/L) and tiamulin (< 0.001–0.125 mg/L). Our results demonstrate the genetic variability among M. hyopneumoniae isolates from pigs of the same farm, with differences in their susceptibility to antimicrobial agents.


Similar content being viewed by others
References
ABPA AB de PA. Relatório Anual 2018.; 2018.
Guimarães D, Amaral G, Maia G, Lemos M, Ito M, Custodio S (2017) Suinocultura: estrutura da cadeia produtiva, panorama do setor no brasil e no mundo e o apoio do BNDES. Agroindústria/ BNDES Setorial 45:85–136
Hillen S, von Berg S, Köhler K, Reinacher M, Willems H, Reiner G (2014) Occurrence and severity of lung lesions in slaughter pigs vaccinated against Mycoplasma hyopneumoniae with different strategies. Prev Vet Med 113(4):580–588. https://doi.org/10.1016/j.prevetmed.2013.12.012
Kuhnert P, Overesch G (2014) Molecular epidemiology of Mycoplasma hyopneumoniae from outbreaks of enzootic pneumonia in domestic pig and the role of wild boar. Vet Microbiol 174(1-2):261–266. https://doi.org/10.1016/j.vetmic.2014.08.022
Yamaguti M (2009) Isolamento de micoplasma de suínos com problemas respiratórios e tipificação dos isolados pela PFGE e seqüenciamento do gene 16S rRNA. Tese, Univ Fed São Paulo, São Paulo- SP, Bras
Simionatto S, Marchioro SB, Maes D, Dellagostin OA (2013) Mycoplasma hyopneumoniae: from disease to vaccine development. Vet Microbiol 165(3-4):234–242. https://doi.org/10.1016/j.vetmic.2013.04.019
Gautier-Bouchardon AV (2018) Antimicrobial Resistance in Mycoplasma spp. Microbiol Spectr 6:1–21. https://doi.org/10.1128/microbiolspec.ARBA-0030-2018
Cook BS, Beddow JG, Manso-Silván L, Maglennon GA, Rycroft AN (2016) Selective medium for culture of Mycoplasma hyopneumoniae. Vet Microbiol 195:158–164. https://doi.org/10.1016/j.vetmic.2016.09.022
Kobisch M, Friis NF (1996) Swine mycoplasmoses. Rev Sci Tech 15(4):1569–1605. https://doi.org/10.1021/pr201115v
Sambrook J, Russel DW (2001) Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Mattsson JG, Bergström K, Wallgren P, Johansson KE, Wallgren PER (1995) Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene;33(4)
Stakenborg T, Vicca J, Butaye P, Imberechts H, Peeters J, de Kruif A, Haesebrouck F, Maes D (2006) A multiplex PCR to identify porcine mycoplasmas present in broth cultures. Vet Res Commun 30(3):239–247. https://doi.org/10.1007/s11259-006-3226-3
Stemke GW (1997) Gene amplification (PCR) to detect and differentiate mycoplasmas in porcine mycoplasmal pneumonia. Lett Appl Microbiol 25(5):327–330. https://doi.org/10.1046/j.1472-765X.1997.00243.x
Assao VS (2017) Detecção e variabilidade genética de mycoplasma hyopneumoniae em amostras de pulmão do estado de Minas Gerais, Brasil. Diss mestrado, Univ Fed Viçosa, Viçosa- MG, Bras. http://www.albayan.ae.
Moreira TS (2016) Perfil genético e epidemiologia molecular de Mycoplasma hyopneumoniae no estado de Minas Gerais, Brasil. Diss mestrado, Univ Fed Viçosa, Viçosa- MG, Bras
Klein U, de Jong A, Moyaert H et al (2017) Antimicrobial susceptibility monitoring of Mycoplasma hyopneumoniae and Mycoplasma bovis isolated in Europe. Vet Microbiol 204(February):188–193. https://doi.org/10.1016/j.vetmic.2017.04.012
Hannan PCT (2000) Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. Vet Res 31(4):373–395. https://doi.org/10.1051/vetres:2000100
Lim K, Furuta Y, Kobayashi I (2012) Large variations in bacterial ribosomal RNA genes. Mol Biol Evol 29(10):2937–2948. https://doi.org/10.1093/molbev/mss101
Dubosson CR, Conzelmann C, Miserez R, Boerlin P, Frey J, Zimmermann W, Häni H, Kuhnert P (2004) Development of two real-time PCR assays for the detection of Mycoplasma hyopneumoniae in clinical samples. Vet Microbiol 102(1-2):55–65. https://doi.org/10.1016/j.vetmic.2004.05.007
Stakenborg T, Vicca J, Butaye P, Maes D, Peeters J, de Kruif A, Haesebrouck F (2005) The diversity of Mycoplasma hyopneumoniae within and between herds using pulsed-field gel electrophoresis. Vet Microbiol 109(1-2):29–36. https://doi.org/10.1016/j.vetmic.2005.05.005
Nicolás MF, Barcellos FG, Hess PN, Hungria M (2007) ABC transporters in Mycoplasma hyopneumoniae and Mycoplasma synoviae: insights into evolution and pathogenicity. Genet Mol Biol 30(SUPPL. 1):202–211. https://doi.org/10.1002/hrm.20267
Reolon LA, Martello CL, Schrank IS, Ferreira HB (2014) Survey of surface proteins from the pathogenic mycoplasma hyopneumoniae strain 7448 using a biotin cell surface labeling approach. PLoS One;9(11). doi:https://doi.org/10.1371/journal.pone.0112596.
Virginio VG, Gonchoroski T, Paes JA, Schuck DC, Zaha A, Ferreira HB (2014) Immune responses elicited by Mycoplasma hyopneumoniae recombinant antigens and DNA constructs with potential for use in vaccination against porcine enzootic pneumonia. Vaccine. 32(44):5832–5838. https://doi.org/10.1016/j.vaccine.2014.08.008
Galli V, Simionatto S, Marchioro SB, Klabunde GHF, Conceição FR, Dellagostin OA (2013) Recombinant secreted antigens from Mycoplasma hyopneumoniae delivered as a cocktail vaccine enhance the immune response of mice. Clin Vaccine Immunol 20(9):1370–1376. https://doi.org/10.1128/CVI.00140-13
Charlebois A, Marois-Créhan C, Hélie P, Gagnon CA, Gottschalk M, Archambault M (2014) Genetic diversity of mycoplasma hyopneumoniae isolates of abattoir pigs. Vet Microbiol 168(2-4):348–356. https://doi.org/10.1016/j.vetmic.2013.11.006
Felde O, Kreizinger Z, Sulyok KM, et al. (2018) Antibiotic susceptibility testing of Mycoplasma hyopneumoniae field isolates from Central Europe for fifteen antibiotics by microbroth dilution method. PLoS One.:1-13. doi:https://doi.org/10.1371/journal.pone.0209030.
Tavío MM, Poveda C, Assunção P, Ramírez AS, Poveda JB (2014) In vitro activity of tylvalosin against Spanish field strains of Mycoplasma hyopneumoniae. Vet Rec 175(21):539. https://doi.org/10.1136/vr.102458
Vicca J, Stakenborg T, Maes D, Butaye P, Peeters J, de Kruif A, Haesebrouck F (2004) In vitro susceptibilities of Mycoplasma hyopneumoniae field isolates. Antimicrob Agents Chemother 48(11):4470–4472. https://doi.org/10.1128/AAC.48.11.4470-4472.2004
Thongkamkoon P, Narongsak W, Kobayashi H, Pathanasophon P, Kishima M, Yamamoto K (2013) In vitro susceptibility of Mycoplasma hyopneumoniae field isolates and occurrence of fluoroquinolone, macrolides and lincomycin resistance. J Vet Med Sci 75(8):1067–1070. https://doi.org/10.1292/jvms.12-0520
Tanner AC, Erickson BZ, Ross RF (1993) Adaptation of the Sensititre® broth microdilution technique to antimicrobial susceptibility testing of Mycoplasma hyopneumoniae. Vet Microbiol 36(3-4):301–306. https://doi.org/10.1016/0378-1135(93)90096-P
Acknowledgments
We would like to thank EMBRAPA, who gently ceded the Mycoplasma hyopneumoniae strain J and Dr. Matheus Gandra Campos for assistance with the statistical analyses. We thank Thiago Augusto Teles de Souza and Wendel Mayer Rezende for the technical support in the analyses.
Availability of data and materials
There is supplementary material.
Funding
We thank the Brazilian Government Agencies. This research was funded by the Coordination for the Improvement of Higher Education Personnel – CAPES (Ph.D. fellowship), the National Council for Scientific and Technological Development (Grant: CNPq304727/2016-4) and the Foundation for Research Support of the State of Minas Gerais (Grants: FAPEMIG-PPM-006618-17 and FAPEMIG-APQ-01327-14).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Project approved by CEUA-UFV under process n°78/2016.
Statement of informed consent
All authors read and agreed to the final manuscript.
Additional information
Responsible Editor: Miliane Moreira Soares de Souza.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Gonzaga, N.F., de Souza, L.F.L., Santos, M.R. et al. Antimicrobial susceptibility and genetic profile of Mycoplasma hyopneumoniae isolates from Brazil. Braz J Microbiol 51, 377–384 (2020). https://doi.org/10.1007/s42770-019-00185-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00185-0