Abstract
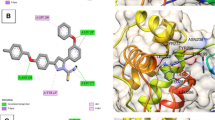
In this study, we established a QSAR model for studying the antiviral activity of substituted thienopyrimidines derivatives as HCV NS3/4A protease inhibitors. We engaged in random analysis to split the datasets. Statistically, a robust model was generated with R2, Q2, and R2pred values of 0.738, 0.637, and 0.692 respectively. The dependability of these models was verified by appropriate testing limits, and this model also met the Golbraikh and Tropsha standard model conditions. The data derived from the established model was employed in suggesting some promising inhibitors of HCV NS3/4A protease and the designed ligand were found to be excellently fixed when anchored with the target and it has the least binding energy of − 197.8 kcal/mol compared to the binding energy of reference ligand (Voxilaprevir) which is − 159.4 kcal/mol. Our analysis indicates that the designed molecules possess the required drug-likeness, bioavailability, synthetic accessibility, and ADMET features.







Similar content being viewed by others
References
Jia S, Zhou W, Wu J, Liu X, Guo S, Zhang J, Zhang X (2020) A biomolecular network-based strategy deciphers the underlying molecular mechanisms of Bupleuri Radix/Curcumae Radix medicine pair in the treatment of hepatitis C. Eur J Integ Med 33:101043
El-Kassem LA, Hawas UW, El-Souda S, Ahmed EF, El-Khateeb W, Fayad W (2019) Anti-HCV protease potential of endophytic fungi and cytotoxic activity. Biocatal Agric Biotechnol 19:101170
González-Grande R, Jiménez-Pérez M, Arjona CG, Torres JM (2016) New approaches in the treatment of hepatitis C. World J Gastroenterol 22(4):1421
Kucherenko A, Pampukha V, Romanchuk KY, Chernushyn SY, Bobrova I, Moroz L, Livshits L (2016) IFNL4 polymorphism as a predictor of chronic hepatitis C treatment efficiency in Ukrainian patients. Cytol Genet 50(5):330–333
World Health Organization (2018) Guidelines for the care and treatment of persons diagnosed with chronic hepatitis C virus infection. Geneva. Licence: CC BY-NC-SA 3.0 IGO. WHO, Geneva
Chahine EB, Sucher AJ, Hemstreet BA (2017) Sofosbuvir/velpatasvir: the first pangenotypic direct-acting antiviral combination for hepatitis C. Ann Pharmacother 51(1):44–53
Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C (2016) Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol 22(34):7824
Coppola N, Alessio L, Onorato L, Sagnelli C, Macera M, Sagnelli E, Pisaturo M (2019) Epidemiology and management of hepatitis C virus infections in immigrant populations. Infect Dis Poverty 8(1):17
Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P (2014) Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59(1):318–327
Liu M, Xu Q, Guo S, Zuo R, Hong Y, Luo Y, Liu Y (2018) Design, synthesis, and structure-activity relationships of novel imidazo [4, 5-c] pyridine derivatives as potent non-nucleoside inhibitors of hepatitis C virus NS5B. Bioorg Med Chem 26(9):2621–2631
Poordad F, McCone J Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Boparai N (2011) Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med 364(13):1195–1206
Bidell MR, McLaughlin M, Faragon J, Morse C, Patel N (2016) Desirable characteristics of hepatitis C treatment regimens: a review of what we have and what we need. Infect Dis Therapy 5(3):299–312
Brown DJ (1984). In: Katritzky AR, Rees CW (eds) Comprehensive Heterocyclic chemistry. Pergamon Press, Oxford
Arthur DE, Ejeh S, Uzairu A (2020) Quantitative structure-activity relationship (QSAR) and design of novel ligands that demonstrate high potency and target selectivity as protein tyrosine phosphatase 1B (PTP 1B) inhibitors as an effective strategy used to model anti-diabetic agents. J Recept Signal Transduct 40:501–520
Neves BJ, Braga RC, Melo-Filho CC, Moreira Filho JT, Muratov EN, Andrade CH (2018) QSAR-based virtual screening: advances and applications in drug discovery. Front Pharmacol 9:1275
Arthur DE, Uzairu A, Mamza P, Abechi S (2016) Quantitative structure–activity relationship study on potent anticancer compounds against MOLT-4 and P388 leukemia cell lines. J Adv Res 7(5):823–837
Roy K, Mitra I, Kar S, Ojha PK, Das RN, Kabir H (2012) Comparative studies on some metrics for external validation of QSPR models. J Chem Inf Model 52(2):396–408
Therese PJ, Manvar D, Kondepudi S, Battu MB, Sriram D, Basu A, Kaushik-Basu N (2014) Multiple e-pharmacophore modeling, 3D-QSAR, and high-throughput virtual screening of hepatitis C virus NS5B polymerase inhibitors. J Chem Inf Model 54(2):539–552
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32(7):1466–1474
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol Inf 29(6–7):476–488
Rogers D (1997) Evolutionary statistics: using a genetic algorithm and model reduction to isolate alternate statistical hypotheses of experimental data. In: Paper presented at the ICGA
Yan F, Liu T, Jia Q, Wang Q (2019) Multiple toxicity endpoint–structure relationships for substituted phenols and anilines. Sci Total Environ 663:560–567
Liu T, Yan F, Jia Q, Wang Q (2020) Norm index-based QSAR models for acute toxicity of organic compounds toward zebrafish embryo. Ecotoxicol Environ Saf 203:110946
Eriksson L, Jaworska J, Worth AP, Cronin MT, McDowell RM, Gramatica P (2003) Methods for reliability and uncertainty assessment and for applicability evaluations of classification-and regression-based QSARs. Environ Health Perspect 111(10):1361–1375
Li F, Li X, Liu X, Zhang L, You L, Zhao J, Wu H (2011) Docking and 3D-QSAR studies on the Ah receptor binding affinities of polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs) and dibenzofurans (PCDFs). Environ Toxicol Pharmacol 32(3):478–485
Evans DA (2014) History of the Harvard ChemDraw project. Angew Chem Int Ed 53(42):11140–11145
Li Z, Wan H, Shi Y, Ouyang P (2004) Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J Chem Inf Comput Sci 44(5):1886–1890
Umar AB, Uzairu A, Shallangwa GA, Uba S (2020) Docking-based strategy to design novel flavone-based arylamides as potent V600E-BRAF inhibitors with prediction of their drug-likeness and ADMET properties. Bull Natl Res Centre 44(1):1–11
Veerasamy R, Rajak H, Jain A, Sivadasan S, Varghese CP, Agrawal RK (2011) Validation of QSAR models-strategies and importance. Int J Drug Des Discov 3:511–519
Danishuddin M, Khan SN, Khan AU (2010) Molecular interactions between mitochondrial membrane proteins and the C-terminal domain of PB1-F2: an in silico approach. J Mol Model 16(3):535–541
Pires DE, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58(9):4066–4072
Funding
No fund received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ejeh, S., Uzairu, A., Shallangwa, G.A. et al. In Silico Design, Drug-Likeness and ADMET Properties Estimation of Some Substituted Thienopyrimidines as HCV NS3/4A Protease Inhibitors. Chemistry Africa 4, 563–574 (2021). https://doi.org/10.1007/s42250-021-00250-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-021-00250-y