Abstract
Despite the wide availability and usage of Gatan’s DigitalMicrograph software in the electron microscopy community for image recording and analysis, nonlinear least-squares fitting in DigitalMicrograph is less straightforward. This work presents a ready-to-use tool, the DMPFIT software package, written in DigitalMicrograph script and C++ language, for nonlinear least-squares fitting of the intensity distribution of atomic columns in atomic-resolution transmission electron microscopy (TEM) images with a general two-dimensional (2D) Gaussian model. Applications of the DMPFIT software are demonstrated both in atomic-resolution conventional coherent TEM (CTEM) images recorded by the negative spherical aberration imaging technique and in high angle annular dark field (HAADF) scanning TEM (STEM) images. The implemented peak-finding algorithm based on the periodicity of 2D lattices enables reliable and convenient atomic-scale metrology as well as intuitive presentation of the resolved atomic structures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Transmission electron microscopes with spherical aberration (\({\mathrm {C}}_{{\rm S}}\)) correction [1,2,3] are capable of resolving individual atoms in materials, and thereby have made it possible to determine positions of individual atoms with the precision of a few picometers. With the sharp increase in the application of aberration-corrected TEM, unprecedented deep insights into materials have been obtained, allowing the connection of material properties with the observed individual constituent atoms [4,5,6,7,8]. It has been of growing interest not only to measure the position of individual atomic columns, but also to determine the atom occupancy and the number of atoms in individual columns [9,10,11,12,13,14,15].
In atomic-resolution images recorded by \({\mathrm {C}}_{{\rm S}}\)-corrected transmission electron microscopes, the intensity distribution of atomic columns, i.e., contrast of atoms, often appears to be peaks centered at the respective column positions. The intensity distribution of the atomic columns have often been phenomenally modeled with a general 2D Gaussian peak [14,15,16,17,18,19,20]. The parameters of Gaussian peaks can be estimated by a nonlinear least-squares fitting (NL-LSQF) method. This allows convenient and precise quantification of atomic column positions. The quantification needs to be conducted necessarily, but often neglected, together with image simulation in order to exclude possible artifacts resulting from the effects of parameters of the instrument (e.g., aberrations) and the sample (e.g., tilting, thickness).
From the physics of scattering point of view, the electron scattering factor \(f_{e}(q)\) at large and small angles can be appropriately parameterized by an analytical formula [21]:
where \(q={{\sin }}(\alpha )/\lambda\), \(\lambda\) is wavelength of the incident electrons, and \(\alpha\) is the scattering semiangle, \(N_{L}\) and \(N_{G}\) are the number of Lorenzians (first summation) and Gaussians (second summation), respectively. Therefore, \(f_{e}(q)\) is predominated by Gaussians (second summation) at low scattering angles and by Lorenzians at high angles.
It is a realistic fact that most microscopes installed in laboratories are equipped with Gatan’s CCD cameras, which are bundled with Gatan’s DigitalMicrograph (DM) for image recording and analysis [22]. However, application of NL-LSQF in DM is not implemented. This compels users to employ alternative software programs like MATLAB, IDL, and Igor Pro for peak finding and fitting in the quantification of atomic-resolution CTEMFootnote 1 and STEM images [14,15,16]. To use these programs, users need extra efforts to import images that they originally recorded using DM to the software and to gain extra knowledge and skills in the use of the software. This, however, is not the case for DM since users have already gotten familiar with the use of DM for recording TEM images before they use it to process and analyze the images. Implementation of NL-LSQF in DM may bring convenience and enable to reduce knowledge and skill barriers in TEM image quantification for the majority of users. Therefore, there is a demand for a software package running within DM for fitting the atomic columns with the 2D Gaussian model for convenient usage.
Fortunately, a free license for the DM software development kit (SDK) can be obtained via online request [23]. The DM-SDK allows users to write the computationally intensive modules in C++, C, and even Fortran, and to write the graphical user interface (GUI) in DM script language to extend the DM’s capabilities. In this paper, we describe a software package called DMPFIT, which enables finding and fitting atomic columns with general 2D Gaussian peaks within DM for facilitating quantitative atomic-resolution CTEM and STEM studies. For the DMPFIT software package, MPFIT C package [24], a MINPACK-1 least-squares fitting library in C, was used as the optimization engine for NL-LSQF, while the peak-finding algorithm and the GUI were written in DM script language.
2 MPFIT
MPFIT is a translation of the MINPACK-1 software package, a particular implementation of Levenberg–Marquardt algorithm to solve least-squares problems originally written in Fortran [24, 25]. MPFIT inherits the merits of being robust, self-contained, and general from the MINPACK-1 package. Moreover, MPFIT enhances its usefulness and convenience to allow convenient parameter constraints setting, Jacobian calculation, and improved capability in calculating the covariance matrix of fitting parameters. Furthermore, MPFIT is provided with extensive documentation including examples of usage and a C version package. It is therefore quite straightforward and easy to port MPFIT as a library to DM-SDK without any modifications. Readers interested in more details on MPFIT are referred to [24].
3 DMPFIT
The DMPFIT software package was written in C++ and DM script languages: C++ was used to define the general 2D Gaussian peak model to perform NL-LSQF by calling MPFIT, and to return the optimized set of fitting parameters to DM; whereas DM script language was used to provide a GUI within DM and to find peaks in the image, and thereby provide initial values for the fitting parameters.
3.1 General 2D Gaussian Model
The general 2D Gaussian peak model implemented in the DMPFIT software package can be described according to Eq. (2).
Here, BG represents constant background, \(I_{0}\) the peak height, \(x_{0}\) and \(y_{0}\) the respective peak position in x and y dimensions, A, B, and C parameters related to the peak width and ellipticity (due to misorientation from the zone axis, residue astigmatism, and so forth). The Gaussian peak is presumed to be non-negative. For each peak, fitting parameters BG, A, B, C, \(x_{0}\), \(y_{0}\), and \(I_{0}\) are optimized by NL-LSQF with MPFIT so that the sum of the weighted squared differences (\({\text {dev}}_{\text {wt}}^2\)) between the data and the model is minimized.
Here, Err denotes the error in the measurements of \(I_{{\exp }}\), which is generally unknown at each pixel in a TEM image. As a consequence of electron counting, Poisson distributed shot-noise depends on the beam intensity, and thereby tends to dominate the noise in electron micrographs recorded by a CCD camera. For more than ten electrons detected, the Poisson distributed shot-noise appears to approach a Gaussian distribution, but the standard deviation of the Gaussian distribution is not constant from pixel to pixel. Instead, it is proportional to the square root of the number of detected electrons in each pixel. For Poisson noise-weighted NL-LSQF, shot-noise is considered to be the main contribution to Err due to its dependence on the number of detected electrons. Err is therefore assumed to be the square root of the image intensity expected according to the model,
i.e., the standard deviation of shot-noise. On the other hand, for uniformly weighted NL-LSQF Err is set to 1. In this case, the least-squares estimators are then identical to the maximum likelihood estimators [20]. Both Poisson noise and uniformly weighted NL-LSQF modes have been implemented in the DMPFIT package and users can select one of the two modes from the GUI.
Because even the atomic columns of the same type may have varied fitting parameters, for instance the peak width, ellipticity, and background, no constraint has been set during the optimization of the fitting parameters. It is possible to implement the software package in a way to optimize all the fitting parameters for all the peaks simultaneously, but with substantially increasing the number of fitting parameters from seven to \(6\cdot N + 1\) (six parameters A, B, C, \(x_{0}\), \(y_{0}\), and \(I_{0}\) for each Gaussian peak plus a constant background parameter BG; N denotes the number of peaks). When the number of peaks increases above 100, however, there will be more than 600 parameters to be optimized, and consequently convergence becomes difficult. Therefore, NL-LSQF is implemented in a peak-wise fashion in the present version of the DMPFIT package.
3.2 Peak-Finding Algorithm
A peak-finding algorithm is used to initialize part of the fitting parameters, specifically including BG, \(x_{0}\), \(y_{0}\), \(I_{0}\), A, and B. The background parameter BG is initialized as the minimum within the defined region, whereas the peak intensity \(I_{0}\) is initialized as the local maximum and its position is used as the initial values for the peak position \(x_{0}\), \(y_{0}\). The initial values for A and B, related to the Gaussian peak width, are provided through the size of the fitting region ((2fR+1)\(^2\) pixels) by fR approximately equal to the full width at half maximum of the Gaussian peak. The ellipticity of the Gaussian peak is initialized as \(C=0\).
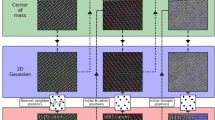
Because the resolved atomic columns in CTEM and STEM images mostly appear periodic, the peak-finding algorithm implemented in DMPFIT is based on the periodicity for the one- and two-dimensional (1D and 2D) modes, which can tolerate moderate deviations from the ideal periodicity. As shown in Fig. 1a, a 2D periodic lattice is defined by the two reference vectors (OA, OB). In the adaptive peak-finding algorithm, an initial peak position is first presumed to be determined by the reference lattices (the center of the square labeled ‘0’ in Fig. 1a). The peak position then is updated as the position where the intensity shows maximum within the presumed fitting region, and thereafter the fitting region is updated to be centered at the new peak position (square labeled ‘1’). Several iterations of the above updating process allow to obtain precise initial values for the fitting parameters, e.g., the constant background BG, the peak intensity \(I_{0}\), and the peak position \(x_{0}\), \(y_{0}\), thereby facilitating the convergence of fitting. This iterative procedure also allows for automatically finding peaks of large deviations from their ideal positions determined by the reference vectors. In practice, five iterations of the updating process appear to be viable.
Moreover, for noisy images, it turns out that peak finding from the low-pass-filtered images gives more reasonable initial values for the fitting parameters compared to those from the unfiltered images. Therefore, a Butterworth filter is used in the peak-finding algorithm. It should be noted here that the raw image is still used as the input for optimizing the fitting parameters by NL-LSQF. Furthermore, a manual mode is implemented to afford users full flexibility for finding non-periodic peaks. Figure 1b, c shows an example for finding peaks in the 2D mode, for which three peaks are selected to define the origin (O) and the two reference vectors (OA, OB), thereby a 2D periodic lattice (marked by green circles) is determined. While for the 1D mode, only OA is used as the reference vector to define a 1D lattice. After a peak has been found by the peak-finding algorithm, DMPFIT will then optimize the fitting parameters for the peak by NL-LSQF.
a Schematic of the adaptive peak-finding algorithm based on the periodicity defined by the two reference vectors OA and OB. Iteratively seeking local maximum and updating the fitting region are used in order to obtain precise initial values of the background and peak intensity and position, thereby allowing finding peaks of some deviations from their ideal positions determined by the reference vectors. The size of the fitting region for each peak is defined as \((2{\mathrm {fR}} + 1)^{2}\). b Experimental atomic-resolution CTEM image of SrTiO\(_{3}\) recorded in the [110] zone axis by the negative spherical aberration (\({\mathrm {C}}_{{\rm S}}\)) imaging (NCSI) technique at 300 kV accelerating voltages with FEI Titan 80–300 kV marked with unit cell structure model: Sr-O: green, Ti: red, O: blue. Image intensity was normalized with the mean of the entire image. Intensity scale: 0.7–1.7. Pixel sampling rate: 0.0091 nm/pixel. Image size: 206\(\times\)200 pixels. c Illustration of 2D peak finding based on periodicity, in which columns labeled O, A, and B define two reference vectors OA and OB, which in turn defines the Sr-O sublattice labeled by green circles
3.3 GUI
Figure 2 shows the GUI of the DMPFIT package. The fR defines the number of pixels used for nonlinear least-squares fitting with the 2D Gaussian model by \((2{\mathrm {fR}} + 1)^2\) pixels. RGB defines the colors for marking the found peaks. The adaptive check box enables users to adaptively find peaks that deviate from the ideal positions in the reference vectors defined lattice. When the Live Update check box is enabled, the peaks will be marked during the finding and fitting process, allowing users to lively monitor whether the peak-finding and fitting process goes well. The ‘smooth’ parameter gives the half-power frequency of the Butterworth filter in percentage of k-space.
For usage in the 2D peak-finding mode, first open an atomic-resolution TEM image, click on the ‘Get Image’ button to create a copy of the front image. Second, click the ‘2D’ button to create the ‘Working’, ‘O’, ‘A’, and ‘B’ ROIs (region of interest) in the front image, and thereafter put ‘O’, ‘A’, and ‘B’ ROIs around the desired peaks from which the lattice of interest shall be defined. Adjust the size and position of the ‘Working’ ROI if necessary; then click the ‘2D’ button again, the peaks within the ‘Working’ ROI defined by the reference lattice will be found and their fitting parameters will be optimized by the NL-LSQF. For each peak, all of the seven optimized fitting parameters, including BG, A, B, C, \(x_{0}\), \(y_{0}\), and \(I_{0}\), as well as their scaled uncertainties with a 95% confidence interval, will be printed in the output window of DM. The scaled uncertainties with a 95% confidence interval are calculated as:
Here, the formal 1-sigma error \(\sigma\) for each optimized fitting parameter is computed from the covariance matrix by MPFIT. \(\chi ^{2}\) is the Chi-square value also returned from the MPFIT library. The degree of freedom \(\nu\) is calculated as the subtraction of the number of the free-fitting parameters (7) from the number of pixels \(({\mathrm {2fR+1}})^2\) for the fitting, and T is the quantile \(Q_{f}({\mathrm {compliment}}(t_{\nu },0.025))\) in which \(t_{\nu }\) represents the Student’s t distribution with \(\nu\) degrees of freedom. Interested readers are referred to Ref. [24] for more details about how the formal 1-sigma errors from the covariance matrix have been calculated by MPFIT for the fitting parameters.
4 Discussion
4.1 Accuracy and Precision
Fitting of the intensity distribution of atomic columns with the general 2D Gaussian model is workable for both CTEM images recorded using the negative spherical aberration (\({\mathrm {C}}_{{\rm S}}\)) imaging (NCSI) technique [26, 27] and HAADF STEM images. The difference between the experimental TEM images and NL-LSQF results reveals that the intensity distributions of atomic columns in both NCSI CTEM and HAADF STEM images fit the general 2D Gaussian model described in Eq. (2) very well. Figure 3a shows an image consisting of Gaussian peaks calculated using the parameters from NL-LSQF results of the NCSI CTEM image shown in Fig. 1b. The difference between the experimental data and the fitting is shown in Fig. 3b, which indicates a nice agreement between the experimental data and the fitting. This is particularly true for Ti and O columns (see Fig. 1a). The doughnut-like dark contrast encircling around the Sr-O columns (smaller dots in Fig. 1b) results from the interference of coherent electrons underlying the image formation. The intensity distribution of image contrast of Sr-O columns appears to be Gaussians only in the region up to the center radius of the doughnut-like dark contrast. It deviates from Gaussians beyond this region giving riseto residue features in the difference image (Fig. 3b).
a Fitted image calculated from the optimized parameters by the nonlinear least-squares fitting of the experimental NCSI CTEM image shown in Fig. 1b. Intensity scale: 0.7–1.7. b Difference between the experimental and the fitted images. Intensity scale: \(-0.13\) to 0.36
The intensity distribution of atomic columns in HAADF STEM images also fits well with the 2D Gaussian model. An image series of 20 experimental HAADF STEM images of SrTiO\(_{3}\) along \(\langle\)001\(\rangle\) zone axis has been recorded and tested. Figure 4a shows the image number ’0’ in the series for instance, and Fig. 4b is the image calculated from the optimized fitting parameters for the intensity distribution of the atomic columns. Since the optimization was performed in a peak-wise fashion, an averaged value of the constant background, \(\bar{B}=\frac{\sum _{i=0}^{N}B_i}{N+1}\), i the peak number, was used in Fig. 4b. As seen in Fig. 4c, the difference between the raw experimental and the fitted images shows a quite featureless background, which can be mainly attributed to random noise and scanning noise.
a An experimental HAADF STEM image of \({{\mathrm{SrTiO}}_3}\) along \(\langle\)001\(\rangle\) zone axis from an image series of 20 images recorded at 200-kV accelerating voltages with an FEI Titan ChemiSTEM microscope. The fast scanning is in the horizontal (i) direction. The image intensity was normalized with the mean of the entire image. The number i and j indicate the numbers of interatomic Sr-Sr distances in x and y directions, respectively. Image size: \(370\times 370\) pixels, pixel sampling rate: 0.0083 nm/pixel. b Fitted image calculated from the optimized fitting parameters of the atomic columns. c Difference image between the experimental and the fitted images
The difference images for the whole image series are shown in Fig. 5a, consistently revealing no systematic deviations of the fitted and experimental images. Moreover, the atomic column distances calculated from the determined atomic positions have been evaluated by taking the Sr-Sr interatomic column distance for instance as marked in Fig. 4a. As shown in Fig. 5b, the mean 95% confidence intervals are ± 6 and ± 9 pm in the fast and slow scanning directions, respectively. These values represent the accuracy of the measurement of atomic distance. For the implemented fitting algorithm, the accuracy for peak position can be down to 0.001 pixel from simulated noise-free Gaussian peaks, which would guarantee sub-pico meter precision of quantification for a typical scale of 0.01 nm/pixel. For experimental data, however, it should be noted that the precision in determination of the atomic column positions and the atomic distances depends on the signal-to-noise ratio and the pixel sampling rate of the recorded images. In addition, lens aberrations and tilts of the electron beam and samples may cause the intensity distribution of atomic columns to deviate from a Gaussian distribution, thereby resulting in poor accuracy and precision. Furthermore, when fitting with raw experimental images, the goodness of fit could be judged by the uncertainties of parameters and the returned \(\chi ^{2}\) for each peak, whereas for fitting with filtered images, one should be aware that the output parameter uncertainties and the obtained \(\chi ^{2}\) will probably not represent the true parameter uncertainties and the true goodness of fit.
a Difference images between the experimental images and the corresponding fitted images calculated from the optimized fitting parameters for the whole image series. For each image, a region as marked in image number ‘0’ was used for the fitting. Intensity scale: \(-0.5\) to 0.5. b Sr-Sr interatomic distances calculated from the optimized fitting parameters by the nonlinear least-squares fitting for the two orthogonal directions as indicated in Fig. 4a. The error bars indicate the 95% confidence intervals obtained from the statistical analysis of the numbered Sr-Sr interatomic distances for the 20 images of the series. The mean 95% confidence intervals are ± 6 pm and ± 9 pm for distances \(i = 0\)–56 and \(j = 0\)–56, respectively
4.2 Peak Intensity Analysis
Using the DMPFIT software package, the integral peak intensity for each column observed in a HAADF STEM image of a \({{\mathrm{SrTi}}_{0.75}{\mathrm{Zr}}_{0.25}{\mathrm O}_3}\) nanocube (Fig. 6a) was obtained by fitting the intensity distribution of the two types of atomic columns with two-dimensional Gaussian functions [28]. The Gaussian peaks from fitting were encoded in green and red colors for Sr and Ti/Zr-O columns, respectively, and overlaid over the HAADF image, which allows the convenient identification of the type of the columns (Fig. 6b). As shown in Fig. 6b, the Sr columns appear to terminate the surfaces of the nanocube at all the four 100 facets. The integral intensities for the Sr atomic columns distribute statistically into a lower intensity range for the columns on the surfaces and a higher intensity range for the inner columns (Fig. 6c and e). In contrast, those for the Ti/Zr-O columns are in a broad and continuous range (Fig. 6d and f), resulting from the inhomogeneity in the number of the Zr atoms that occupy the B-sites in individual columns.
Quantification of the integral intensity of atomic columns from a HAADF STEM image of a \({{\mathrm{SrTi}}_{0.75}{\mathrm{Zr}}_{0.25}{\mathrm O}_3}\) nanocube. a HAADF STEM image. b HAADF STEM image superposed with fitted color-scale two-dimensional Gaussian peaks of atomic columns. c, d Integral peak intensity of the Sr and Ti/Zr-O columns, respectively. Arrows in d indicate Zr-rich columns showing exceptional brightness. e, f Histograms of the integral intensities of the Sr and Ti/Zr-O columns, respectively (reproduced from Ref. [28] with permission from American Chemical Society)
The peak-finding algorithm implemented in DMPFIT based on the periodicity shows particular advantages in finding peaks of atom columns within different sublattices. Each sublattice can be straightforwardly labeled in a specified color using quantified positions of atom columns (Fig. 7a). On the other hand, users may create colored composite images using images of Gaussian peaks calculated from the optimized fitting parameters for intuitively presenting data (Fig. 7b). Using the thresholding method to find peaks restricted to a given sublattice will fail if the intensity difference between atomic columns in different sublattices is small.
Intuitive presentation of the resolved atomic structure in Fig. 1a. Sr-O: green, Ti. red, O: blue. a Atomic columns labeled by marks drawn using the optimized peak positions. b Atomic columns labeled by colored composite image using the Gaussian peaks calculated from the optimized fitting parameters
4.3 Peak Position Analysis
The DMPFIT package facilitates the peak position analysis (PPA) of atomic contrast of CTEM and STEM images and thereby allows straightforward mapping of strain fields of materials at the atomic scale. Figure 8 shows a comparison of the geometry phase analysis (GPA) and PPA methods for strain mapping from a CTEM image (Fig. 8a) of a cross-section sample of Fe-doped \({{\mathrm{SrTiO}}_3}\) film. The image was recorded along the \(\langle 001\rangle\) direction using the NCSI technique. In the image (Fig. 8a, inset), small bright dots correspond to Sr and Ti-O columns showing about the same brightness in contrast, and are therefore not distinguishable from one another. The oxygen atom columns show lower contrast, which appear to be more delocalized than the Sr and Ti-O columns. Antiphase domains or even clusters have been observed in this Fe-doped \({{\mathrm{SrTiO}}_3}\) film [29]. The location of an antiphase domain is indicated by a white dashed line square in the CTEM image (Fig. 8a). Axial strain maps \(\varepsilon _{{xx}}\) and \(\varepsilon _{{yy}}\) shown in Fig. 8b and c were calculated from the positions of the Sr and Ti-O columns by the PPA. The measured strain maps reveal that the lattices show tensile strains in the domains but compressive strains at the antiphase boundaries, where \({{\mathrm{TiO}}_6}\) octahedra appear to be sharing edges instead of corner [29].
Strain mapping from atomic-resolution CTEM image. a CTEM image of an Fe-doped \({{\mathrm{SrTiO}}_3}\) film recorded along the \(\langle 001\rangle\) direction using the NCSI technique. The inset shows a magnified region. The dashed line square shows the location of an antiphase nanodomain [29]. b, c Axial strain maps \(\varepsilon _{{xx}}\) and \(\varepsilon _{{yy}}\) calculated using the peak position analysis (PPA) methods, and e, f using the geometry phase analysis (GPA). d The central part of the diffractogram of the image shown in Fig. 8a, in which the two {220} reflections labeled as 1 and 2 were chosen for the GPA. The cut-off of the reflections is marked with red and green circles. The location of antiphase nanodomain is also indicated by a dashed line square in the strain maps
The GPA [30, 31] makes use of a pair of noncolinear reflections with reference to the central spot in the Fourier transformation of the image. Compared to the real space PPA for mapping strains, the GPA appears to be more sensitive to noise and choice of the pair of reflections. Strain maps obtained from GPA using different pairs of reflections may lead to results that are substantially different from one another. We performed the GPA using homemade software [32, 33]. For the \(\varepsilon _{{xx}}\) and \(\varepsilon _{{yy}}\) strain maps shown in Fig. 8e and f, two {220} reflections were used. The selected reflections were smoothed with a simple cosine low-pass filter that has a measure of 1 at the center of the reflections and is approaching through the cosine function to 0 at the cut-off marked by the green and red circles (Fig. 8d). Using these exact parameters, the strain maps obtained using the GPA method (Fig. 8e and f) quantitatively match those obtained using the PPA method (Fig. 8b and c).
On the other hand, precise quantification column positions in atomic-resolution TEM images allows to infer, unit cell by unit cell, the local polarization resulting from a noncentrosymmetric distribution of cations and anions in ferroelectric materials [4, 5]. Figure 9a shows a map of local spontaneous polarization vectors calculated from the measured peak positions of Hf (yellow dots) and O (blue dots) columns in an NCSI TEM image of a \({{\mathrm{HfO}}_2}\) nanocrystal using the DMPFIT software package. The polarization in the \({{\mathrm{HfO}}_2}\) nanocrystals results from twinning, through which a metastable polar orthorhombic phase having space group \(Pbc2_1\) is formed at the twin boundary [4]. The polarization is essentially aligned along the vertical direction. In the vertical blocks labeled with 3 to 5 and 9 in Fig. 9a, the averaged values for the vertical component of the spontaneous polarization \(P_{{y}}\) is in the range of 30–40 \(\upmu {\mathrm {C}}\,{\mathrm {cm}}^{-2}\) (Fig. 9b). These values are in good agreement with those (filled circles in Fig. 9b) calculated using atomic positions from the refined structure via image matching between the experiment and simulation. As the magnitude of specimen tilt is small (x: \(-0.25\) mrad, y: \(-0.62\) mrad), the agreement can be explained by the about 2-nm-thin thickness of the specimen, which mitigated the effects of beam tilt (x: \(-73\) nm, y: \(-12.5\) nm).Footnote 2 Moreover, the carbon support resulted in considerable fluctuations in the image contrast, which unavoidably caused extra errors in the measurement by peak fitting. These errors might overwhelm the effect resulting from beam tilt. Nevertheless, we wish to point out that special care needs to be taken for beam and specimen tilts in the imaging experiments, which may result in artifacts from the peak positions of the intensity distribution of atomic contrast [34]. In particular for specimens of compounds, beam and specimen tilts as well as a combination of them may affect the intensity distribution of image contrast of lighter and heavier atomic columns in different extents [34]. As a result, peak positions of the intensity distribution of atomic contrast do not accurately correspond to the true column positions of the materials. Artifacts resulting from the non-negligible beam and specimen tilts may thus be possibly misinterpreted as physical properties, e.g., polarization.
Polarization in \({\mathrm{HfO}_2}\) nanocrystals. a Map of local spontaneous polarization vectors (arrows) calculated using the measured peak positions of Hf (yellow dots) and O (blue dots) superposed on the NCSI image. The length of the arrows represents the modulus of the polarization vectors with respect to the yellow scale bar in the upper left corner. The arrowheads point in the polarization directions. Horizontal dimension is of 50 nm. b Statistical analysis of vertical (\(P_{{y}}\), upper panel) and horizontal (\(P_{{x}}\), lower panel) components of the spontaneous polarization (\(\varvec{P}_{{\rm s}}\)) for structures in each vertical block. The bars represent the average values (yellow filled for upward and cyan filled for downward). The error bar represents the standard deviation with respect to the averaged value for each block. The solid red circles denote the values calculated using the atomic positions of the optimized structure by experiment-simulation matching (reproduced from Ref. [4] with permission from Elsevier)
Experiment-simulation matching seeks optimized parameters of the microscope and specimen, including positions of all the atoms, at which the simulated image matches best with the experimental image. This allows separating artifacts resulting from non-negligible beam and specimen tilts. Experiment-simulation matching by an optimization scheme is viable for CTEM images but not practical for STEM images. Simulation using the multislice method for a CTEM image takes only one beam to transmit and propagate through thin slices of the specimen. Simulation for a STEM image, however, needs to repeat the beam transmission and propagation through all the slices at every pixel position, and thus, can be thousands or millions of times slower than that for a CTEM image. Nevertheless, a number of deliberate tilts of beam and specimen can be applied in a limited number of STEM image simulations to estimate the influence of parameters of specimen and microscope and hence the errors in the measurement of atom positions by peak fitting.
5 Conclusions
In conclusion, a software package, DMPFIT, has been successfully developed. It runs within DM for quantification of atomic columns in atomic-resolution (S)TEM images by NL-LSQF with a general 2D Gaussian model. DMPFIT uses the MPFIT C package, a MINPACK-1 least-squares fitting library in C, as the optimization engine for NL-LSQF. The implemented peak-finding algorithm based on 2D periodicity has been found to have particular advantages in quantification of physical properties associated with sublattices. Applications of the DMPFIT software package have been demonstrated by quantification of the intensity and position of columns observed in experimental NCSI STEM and HAADF STEM images. Based on the peak position analyses, properties such as strains and ferroelectric polarization can be quantified on the atomic scale. The DMPFIT software package works as a plugin to Gatan’s DigitalMicrograph software, also known as Gatan Microscopy Suite (GMS), with version 2. As a ready-to-use tool for atomic-scale metrology by peak finding and fitting in atomic-resolution TEM images, the DMPFIT software package is freely available via the Internet after the publication of the manuscript.
Change history
23 November 2022
A Correction to this paper has been published: https://doi.org/10.1007/s41871-022-00159-1
Notes
Atomic-resolution CTEM is a special case of high-resolution TEM (HRTEM) for which contrast of atoms is resolved. However, at many imaging conditions HRTEM resolves contrast of only lattice fringes but not atoms. In these cases, HRTEM and atomic-resolution CTEM are not equivalent.
These parameters, along with other parameters about the specimen and the microscope, were estimated by experiment-simulation matching. They can be found in Table S1 in the Supplemental Information of Ref. [4].
References
Haider M, Uhlemann S, Schwan E, Rose H, Urban B, Kabius K (1998) Electron microscopy image enhanced. Nature 392(6678):768–769. https://doi.org/10.1038/33823
Uhlemann S, Haider M (1998) Residual wave aberrations in the first spherical aberration corrected transmission electron microscope. Ultramicroscopy 72(3):109–119. https://doi.org/10.1016/S0304-3991(97)00102-2
Dellby N, Krivanek OL, Nellist PD, Batson PE, Lupini AR (2001) Progress in aberration-corrected scanning transmission electron microscopy. J Electron Microsc 50(3):177–185. https://doi.org/10.1093/jmicro/50.3.177
Du H, Groh C, Jia CL, Ohlerth T, Dunin-Borkowski RE, Simon U, Mayer J (2021) Multiple polarization orders in individual twinned colloidal nanocrystals of centrosymmetric HfO2. Matter 4(3):986–1000. https://doi.org/10.1016/j.matt.2020.12.008
Lu L, Nahas Y, Liu M, Du H, Jiang Z, Ren S, Wang D, Jin L, Prokhorenko S, Jia CL, Bellaiche L (2018) Topological defects with distinct dipole configurations in PbTiO\(_{3}\)/SrTiO\(_{3}\) multilayer films. Phys Rev Lett 120(17):177601. https://doi.org/10.1103/PhysRevLett.120.177601
Jia CL, Mi SB, Urban K, Vrejoiu I, Alexe M, Hesse D (2008) Atomic-scale study of electric dipoles near charged and uncharged domain walls in ferroelectric films. Nat Mater 7(1):57–61. https://doi.org/10.1038/nmat2080
Jia C, Mi S, Urban K, Vrejoiu I, Alexe M, Hesse D (2009) Effect of a single dislocation in a heterostructure layer on the local polarization of a ferroelectric layer. Phys Rev Lett 102(11):117601. https://doi.org/10.1103/PhysRevLett.102.117601
Jia CL, Jin L, Wang D, Mi SB, Alexe M, Hesse D, Reichlova H, Marti X, Bellaiche L, Urban KW (2015) Nanodomains and nanometer-scale disorder in multiferroic bismuth ferrite single crystals. Acta Mater 82:356–368. https://doi.org/10.1016/j.actamat.2014.09.003
Jia CL, Urban K, Lentzen M (2003) Atomic-resolution imaging of oxygen in perovskite ceramics. Science 299(5608):870–873. https://doi.org/10.1126/science.1079121
Jia CL, Urban K (2004) Atomic-resolution measurement of oxygen concentration in oxide materials. Science 303(5666):2001–2004. https://doi.org/10.1126/science.1093617
Jia CL, Urban KW, Alexe M, Hesse D, Vrejoiu I (2011) Direct observation of continuous electric dipole rotation in flux-closure domains in ferroelectric Pb(Zr, Ti)O\(_{3}\). Science 331(6023):1420–1423. https://doi.org/10.1126/science.1200605
Jia CL, Mi SB, Barthel J, Wang DW, Dunin-Borkowski RE, Urban KW, Thust A (2014) Determination of the 3D shape of a nanoscale crystal with atomic resolution from a single image. Nat Mater 13(11):1044–1049. https://doi.org/10.1038/nmat4087
De Backer A, Martinez G, Rosenauer A, Van Aert S (2013) Atom counting in HAADF STEM using a statistical model-based approach: methodology, possibilities, and inherent limitations. Ultramicroscopy 134:23–33. https://doi.org/10.1016/j.ultramic.2013.05.003
LeBeau JM, Findlay SD, Allen LJ, Stemmer S (2010) Standardless atom counting in scanning transmission electron microscopy. Nano Lett 10(11):4405–4408. https://doi.org/10.1021/nl102025s
Yankovich AB, Berkels B, Dahmen W, Binev P, Sanchez SI, Bradley SA, Li A, Szlufarska I, Voyles PM (2014) Picometre-precision analysis of scanning transmission electron microscopy images of platinum nanocatalysts. Nat Commun 5:5155. https://doi.org/10.1038/ncomms5155
Houben L (2022) iMtools. https://er-c.org/index.php/software/stem-data-analysis/imtools/. Accessed 10 Jan 2022
den Dekker A, Gonnissen J, De Backer A, Sijbers J, Van Aert S (2013) Estimation of unknown structure parameters from high-resolution (S)TEM images: what are the limits? Ultramicroscopy 134:34–43. https://doi.org/10.1016/j.ultramic.2013.05.017
Van Aert S, Verbeeck J, Erni R, Bals S, Luysberg M, Dyck DV, Tendeloo GV (2009) Quantitative atomic-resolution mapping using high-angle annular dark field scanning transmission electron microscopy. Ultramicroscopy 109(10):1236–1244. https://doi.org/10.1016/j.ultramic.2009.05.010
den Dekker A, Van Aert S, van den Bos A, Van Dyck D (2005) Maximum likelihood estimation of structure parameters from high-resolution electron microscopy images. Part I: A theoretical framework. Ultramicroscopy 104(2):83–106. https://doi.org/10.1016/j.ultramic.2005.03.001
Van Aert S, den Dekker A, van den Bos A, Van Dyck D, Chen J (2005) Maximum likelihood estimation of structure parameters from high-resolution electron microscopy images. Part II: A practical example. Ultramicroscopy 104(2):107–125. https://doi.org/10.1016/j.ultramic.2005.03.002
Kirkland EJ (2020) Advanced computing in electron microscopy, 3rd edn. Springer, Cham. https://doi.org/10.1007/978-3-030-33260-0 (Accessed 17 Jul 2020)
Gatan Inc. DigitalMicrograph\(^{{\rm TM}}\). http://www.gatan.com/products/tem-analysis/gatan-microscopy-suite-software (2015). Accessed 25 Jun 2015
Gatan Inc. DigitalMicrograph\(^{{\rm TM}}\) 2.x SDK. http://www.gatan.com/resources/scripts (2015). Accessed 25 Jun 2015
Markwardt CB (2022) Non-linear least squares fitting in IDL with MPFIT. In: Bohlender DA, Durand D Dowler P (eds) Astronomical data analysis software and systems XVIII: proceedings of a workshop held at Hotel Loews Le Concorde, Quebec City, QC, Canada, 2-5 November 2008, vol 411, p 251 (Astronomical Society of the Pacific, 2009). arXiv:0902.2850. Accessed 10 Jan 2022
Moré JJ (2022) The Levenberg–Marquardt algorithm: Implementation and theory. In: Watson GA (ed) Numerical analysis. Springer, Berlin, Heidelberg, 1978, pp 105–116. https://doi.org/10.1007/BFb0067700. Accessed 10 Jan 2022
Jia CL, Lentzen M, Urban K (2004) High-resolution transmission electron microscopy using negative spherical aberration. Microsc Microanal 10(2):174–184. https://doi.org/10.1017/S1431927604040425
Urban KW, Jia CL, Houben L, Lentzen M, Tillmann K, Mi SB (2009) Negative spherical aberration ultrahigh-resolution imaging in corrected transmission electron microscopy. Philos Trans R Soc A: Math Phys Eng Sci 367(1903):3735–3753. https://doi.org/10.1098/rsta.2009.0134
Du H, Jia CL, Mayer J (2016) Surface atomic structure and growth mechanism of monodisperse 1 0 0-faceted strontium titanate zirconate nanocubes. Chem Mater 28(2):650–656. https://doi.org/10.1021/acs.chemmater.5b04486
Du H, Jia CL, Mayer J, Barthel J, Lenser C, Dittmann R (2015) Atomic structure of antiphase nanodomains in Fe-doped SrTiO\(_{{\rm 3}}\) films. Adv Funct Mater 25(40):6369–6373. https://doi.org/10.1002/adfm.201500852
Takeda M, Suzuki J (1996) Crystallographic heterodyne phase detection for highly sensitive lattice-distortion measurements. J Opt Soc Am A 13(7):1495–1500. https://doi.org/10.1364/JOSAA.13.001495
Hÿtch MJ, Snoeck E, Kilaas R (1998) Quantitative measurement of displacement and strain fields from HREM micrographs. Ultramicroscopy 74(3):131–146. https://doi.org/10.1016/S0304-3991(98)00035-7
Du H (2018) GPA—geometrical phase analysis software. https://er-c.org/index.php/software/stem-data-analysis/gpa/
Du H, Jia CL, Houben L, Metlenko V, De Souza RA, Waser R, Mayer J (2015) Atomic structure and chemistry of dislocation cores at low-angle tilt grain boundary in SrTiO\(_{{\rm 3}}\) bicrystals. Acta Mater 89:344–351. https://doi.org/10.1016/j.actamat.2015.02.016
Du H, Jia CL (2022) Epitaxial growth of complex metal oxides. In: Ch. 13 High-resolution transmission electron microscopy and spectroscopy of epitaxial metal oxides, 2nd edn. Elsevier (in production)
Acknowledgements
Financial support from the German Research Foundation (SFB917) is acknowledged. The author thanks Gatan Inc. for providing the free license of the DM SDK.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conficts of interest.
Additional information
The original online version of this article was revised: The declaration text was missing
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Du, H. DMPFIT: A Tool for Atomic-Scale Metrology via Nonlinear Least-Squares Fitting of Peaks in Atomic-Resolution TEM Images. Nanomanuf Metrol 5, 101–111 (2022). https://doi.org/10.1007/s41871-022-00137-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41871-022-00137-7