Abstract
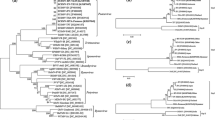
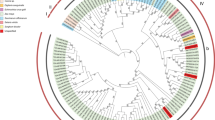
Yellow leaf of sugarcane is caused by Sugarcane yellow leaf virus (SCYLV) and has been a threat to global sugar industry. Understanding the diversity and genetic structure of plant virus populations may help to develop strategies to control virus-induced diseases. In this study, demography, genetic differentiation and selection pressure were assessed based on 57 nucleotide sequences of the P0 (567 nt) and 54 nucleotide sequences of the P1 (1413 nt) genes of SCYLV isolates. Phylogenetic analysis revealed three phylogeographical groups: African (Clade I), Chinese/American (Clade II), and Asian (Clade III). Genetic distance values between groups (0.034–0.250) were higher than within-group values (0.011–0.033). The African group was in a state of expansion, with lower nucleotide sequence diversity. Except for those isolates within the Asian group, SCYLV showed infrequent gene flow within or between phylogenetic groups. Analysis of selection pressure suggested that these two genes have been subjected to negative (purifying) selection. However, some sites, especially in the P0 coding region of the Chinese/American group, have undergone positive selection. Overall, aside from recombination and founder effects, gene flow and purifying selection are two important factors driving SCYLV evolution.

Similar content being viewed by others
References
Abu Ahmad Y, Rassaby L, Royer M, Borg Z, Braithwaite KS, Mirkov TE, Irey M, Perrier X, Smith G, Rott P (2006a) Yellow leaf of sugarcane is caused by at least three different genotypes of Sugarcane yellow leaf virus, one of which predominates on the Island of Reunion. Arch Virol 151:1355–1371
Abu Ahmad Y, Royer M, Daugrois JH, Costet L, Lett JM, Victoria JI, Girard JC, Rott P (2006b) Geographical distribution of four Sugarcane yellow leaf virus genotypes. Plant Dis 90:1156–1160
Chinnaraja C, Viswanathan R, Karuppaiah R, Bagyalakshmi K, Malathi P, Parameswari B (2013) Complete genome characterization of Sugarcane yellow leaf virus from India: evidence for RNA recombination. Eur J Plant Pathol 135:335–349
Daniels J, Roach BT (1987) Taxonomy and evolution. In: Heinz DJ (ed) Sugarcane improvement through breeding. Elsevier, Amsterdam, pp 7–84
Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL (2010) Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457
Desbiez C, Moury B, Lecoq H (2011) The hallmarks of “green” viruses: do plant viruses evolve differently from the others? Infect Genet Evol 11:812–824
Drake JW, Holland JJ (1999) Mutation rates among RNA viruses. Proc Natl Acad Sci U S A 96:13910–13913
ElSayed AI, Weig AR, Komor E (2011) Molecular characterization of Hawaiian Sugarcane yellow virus leaf genotypes and their phylogenetic relationship to strains from other sugarcane-growing countries. Eur J Plant Pathol 129:399–412
ElSayed AI, Boulila M, Komor E, Zhu YJ (2012) Putative recombination signature and significance of insertion/deletion events in the RNA-dependent RNA polymerase coding region of Sugarcane yellow leaf virus. Biochimie 94:1764–1772
ElSayed AI, Boulila M, Rot P (2014) Molecular evolutionary history of Sugarcane yellow leaf virus based on sequence analysis of RNA-dependent RNA polymerase and putative aphid transmission factor-coding genes. J Mol Evol 78:349–365
ElSayed AI, Komor E, Boulila M, Viswanathan R, Odero DC (2015) Biology and management of Sugarcane yellow leaf virus: an historical overview. Arch Virol 160:2921–2934
Fargette D, Pinel A, Abubakar Z, Traoré O, Brugidou C, Fatogoma S, Hébrard E, Choisy M, Séré Y, Fauquet C, Konaté G (2004) Inferring the evolutionary history of Rice yellow mottle virus from genomic, phylogenetic, and phylogeographic studies. J Virol 78:3252–3261
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Gao SJ, Lin YH, Pan YB, Damaj MB, Wang QN, Mirkov TE, Chen RK (2012) Molecular characterization and phylogenetic analysis of Sugarcane yellow leaf virus isolates from China. Virus Genes 45:340–349
Haenni AL (2008) Virus evolution and taxonomy. In: Roossinck MJ (ed) Plant virus evolution. Springer, New York, pp 205–217
Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155:2011–2014
Hudson RR, Boos DD, Kaplan NL (1992) A statistical test for detecting geographic subdivision. Mol Biol Evol 9:138–151
Joomun N, Dookun-Saumtally A (2010) Occurrence of three genotypes of Sugarcane yellow leaf virus in a variety collection in Mauritius. Sugar Tech 12:312–316
Lehrer AT, Wu KK, Komor E (2009) Impact of Sugarcane yellow leaf virus (SCYLV) on growth and sugar yield of sugarcane. J Gen Plant Pathol 75:288–296
Li WH (1993) Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol 36:96–99
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lin YH, Gao SJ, Damaj MB, Fu HY, Chen RK, Mirkov TE (2014a) Genome characterization of Sugarcane yellow leaf virus from China reveals a novel recombinant genotype. Arch Virol 159:1421–1429
Lin YH, Xiao SH, Liu YH, Chen JS, Fu HY, Chen RK, Gao SJ (2014b) Molecular characterization and analysis of suppression of RNA silence by the P0 protein encoded by Sugarcane yellow leaf virus (in Chinese). Sci Agric Sin 47:4627–4636
Lin YH, Wu XB, Liu YH, Xiao SH, Zhou GH, Chen RK, Fu HY, Gao SJ (2015) Molecular detection, genetic diversity and distribution of Sugarcane yellow leaf virus genotypes in China. Trop Plant Pathol 40:200–207
Mangwende T, Wang ML, Borth W, Hu J, Moore PH, Mirkov TE, Albert HH (2009) The P0 gene of Sugarcane yellow leaf virus encodes an RNA silencing suppressor with unique activities. Virology 384:38–50
Martin RR, Keese PK, Young JM, Waterhouse PM, Gerlach WL (1990) Evolution and molecular biology of luteoviruses. Annu Rev Phytopathol 28:341–363
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463
Meirmans PG (2006) Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60:2399–2402
Moonan F, Mirkov TE (2002) Analyses of genotypic diversity among North, South, and Central American isolates of Sugarcane yellow leaf virus: evidence for Colombian origins and for intraspecific spatial phylogenetic variation. J Virol 76:1339–1348
Moonan F, Molina J, Mirkov TE (2000) Sugarcane yellow leaf virus: an emerging virus that has evolved by recombination between luteoviral and poleroviral ancestors. Virology 269:156–171
Moury B, Simon V (2011) dN/dS-based methods detect positive selection linked to trade-offs between different fitness traits in the coat protein of Potato virus Y. Mol Biol Evol 8:2707–2717
Nagy PD (2008) Recombination in plant RNA viruses. In: Roossinck MJ (ed) Plant virus evolution. Springer, New York, pp 133–156
Nickel H, Kawchuk L, Twyman RM, Zimmermann S, Junghans H, Winter S, Fischer R, Prüfer D (2008) Plantibody-mediated inhibition of the Potato leaf roll virus P1 protein reduces virus accumulation. Virus Res 136:140–145
Pagán I, Holmes EC (2010) Long-term evolution of the Luteoviridae: time scale and mode of virus speciation. J Virol 84:6177–6187
Pamilo P, Bianchi NO (1993) Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol 10:271–281
Posada D, Crandall KA (2002) The effect of recombination on the accuracy of phylogeny estimation. J Mol Evol 54:396–402
Rott P, Comstock JC, Croft BJ, Kusalwong A, Saumtally S (2005) Advances and challenges in sugarcane pathology: a review of the 2003 Pathology Workshop. Proc Int Soc Sugar Cane Technol 25:607–614
Rozas J, Sanchez-Del Barrio JC, Messeguer X, Rozas R (2003) DnaSP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Sadowy E, Juszczuk M, David C, Gronenborn B, Hulanicka MD (2001) Mutational analysis of the proteinase function of Potato leafroll virus. J Gen Virol 82:1517–1527
Scagliusi SM, Lockhart BEL (2000) Transmission, characterization, and serology of a luteovirus associated with yellow leaf syndrome of sugarcane. Phytopathology 90:120–124
Schenck S (1990) Yellow leaf syndrome—a new sugarcane disease. Hawaii Sugar Plant Assoc Annu Rep, 38–39
Sérémé D, Lacombe S, Konaté M, Bangratz M, Pinel-Galzi A, Fargette D, Traoré AS, Konaté G, Brugidou C (2014) Sites under positive selection modulate the RNA silencing suppressor activity of Rice yellow mottle virus movement protein P1. J Gen Virol 95:213–218
Sharpe P (1998) Sugar cane: past and present. Ethnobotanical Leaflets 3:6
Smith GR, Borg Z, Lockhart BE, Braithwaite KS, Gibbs MJ (2000) Sugarcane yellow leaf virus: a novel member of the Luteoviridae that probably arose by inter-species recombination. J Gen Plant Pathol 81:1865–1869
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Wang MQ, Zhou GH (2010) A near-complete genome sequence of a distinct isolate of Sugarcane yellow leaf virus from China, representing a sixth new genotype. Virus Genes 41:268–272
Wang MQ, Xu DL, Li R, Zhou GH (2012) Genotype identification and genetic diversity of Sugarcane yellow leaf virus in China. Plant Pathol 61:986–993
Wright S (1951) The genetic structure of populations. Ann Eugenics 15:323–354
Acknowledgments
This research was supported by a grant from the National Natural Science Foundation of China (31170345), an Earmarked Fund from the China Agriculture Research System (CARS-20-2-4), and the Major Science and Technology Project of Fujian Province (2015NZ0002-2) in China.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Section Editor: Alice Inoue-Nagata
Chun-Hui Zhao and Yi-Hua Lin contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhao, CH., Lin, YH., Pan, YB. et al. Population structure and selection pressure analysis among Sugarcane yellow leaf virus isolates based on P0 and P1 sequences. Trop. plant pathol. 41, 237–245 (2016). https://doi.org/10.1007/s40858-016-0096-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-016-0096-2