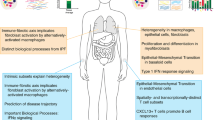
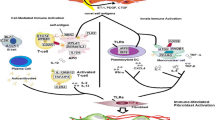
Opinion statement
Purpose of review Systemic sclerosis (SSc) has been difficult to treat due to disease heterogeneity and an incomplete understanding of the molecular mechanisms underlying pathogenesis. Some patients show improvement on existing or experimental therapies while others show no significant response. Due to these challenges, clinical trials often do not meet their primary clinical endpoints. Here we review the data-driven genomewideapproaches that have recently been employed to characterize the molecular changes observed in SSc patients.
Recent findings Understanding an individual patient’s gene expression phenotype or genotype, a cornerstone of personalized medicine, has been proposed to help determine which therapies are most likely to treat their disease. Network and systems biology methods have been applied to the compendium of publicly available SSc data and suggest that there are shared mechanisms driving the disease in different organs. Computational methods have also been applied to meta-analyses of SSc clinical trials. These approaches have led to the prediction of potential combination therapies that target the multiple pathways deregulated in SSc, which can now be tested in the clinic.
Summary Data driven methods to analyze compendia of data are providing additional insights into SSc and related conditions. Modern bioinformatics and systems biology are ecosystems of data and code that are growing exponentially. Data integration allows researchers to combine multiple underpowered studies, a concern in a rare disorder such as SSc, for greater gain.

Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One. 2008;3(7):e2696.
Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012;132(5):1363–73.
Hinchcliff M, Huang CC, Wood TA, Matthew Mahoney J, Martyanov V, Bhattacharyya S, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol. 2013;133(8):1979–89.
Pendergrass SA, Hayes E, Farina G, Lemaire R, Farber HW, Whitfield ML, et al. Limited systemic sclerosis patients with pulmonary arterial hypertension show biomarkers of inflammation and vascular injury. PLoS One. 2010;5(8):e12106.
Risbano MG, Meadows CA, Coldren CD, Jenkins TJ, Edwards MG, Collier D, et al. Altered immune phenotype in peripheral blood cells of patients with scleroderma-associated pulmonary hypertension. Clin Transl Sci. 2010;3(5):210–8.
Cheadle C, Berger AE, Mathai SC, Grigoryev DN, Watkins TN, Sugawara Y, et al. Erythroid-specific transcriptional changes in PBMCs from pulmonary hypertension patients. PLoS One. 2012;7(4):e34951.
Hsu E, Shi H, Jordan RM, Lyons-Weiler J, Pilewski JM, Feghali-Bostwick CA. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheum. 2011;63(3):783–94.
Christmann RB, Sampaio-Barros P, Stifano G, Borges CL, de Carvalho CR, Kairalla R, et al. Association of Interferon- and transforming growth factor β-regulated genes and macrophage activation with systemic sclerosis-related progressive lung fibrosis. Arthritis Rheumatol. 2014;66(3):714–25.
• Taroni JN, Martyanov V, Huang C, Mahoney JM, Hirano I, Shetuni B, et al. Molecular characterization of systemic sclerosis esophageal pathology identifies inflammatory and proliferative signatures. Arthritis Res Ther. 2015;17:194. First report of the intrinsic gene expression subsets first identified in skin in a second organ system in SSc patients.
•• Taroni JN, Greene CS, Martyanov V, Wood TA, Christmann RB, Farber HW, et al. A novel multi-network approach reveals tissue-specific cellular modulators of fibrosis in systemic sclerosis. Genome Med. 2017;9(1):27. This study is the first network analysis of multiple tissues in SSc and finds key similarities in the immune- and fibrotic-related expression patterns across tissues, implicating alternatively activated macrophages in SSc pathogenesis.
Oswald M, Curran ME, Lamberth SL, Townsend RM, Hamilton JD, Chernoff DN, et al. Modular analysis of peripheral blood gene expression in rheumatoid arthritis captures reproducible gene expression changes in tumor necrosis factor responders. Arthritis Rheumatol. 2015;67(2):344–51.
•• Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165(3):551–65. Report of utilizing molecular networks to stratify patients with systemic lupus erythematosus.
Hudson M, Baron M, Colmegna I, Bernatsky S, Klein Oros K, Pastinen T, et al. Novel approaches to discovery of biomarkers in rheumatoid arthritis: comment on the article by Oswald et al. Arthritis Rheum. 2015;67(8):2276–7.
Johnson ME, Mahoney JM, Taroni J, Sargent JL, Marmarelis E, Wu MR, et al. Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts. PLoS One. 2015;10(1):e0114017.
•• Mahoney JM, Taroni J, Martyanov V, Wood TA, Greene CS, Pioli PA, et al. Systems level analysis of systemic sclerosis shows a network of immune and profibrotic pathways connected with genetic polymorphisms. PLoS Comput Biol. 2015;11(1):e1004005. Data integration and network analysis of SSc skin cohorts that shows an interconnection of immune and fibrotic pathways. This study connects the genetic risk polymorphisms for SSc with the inflammatory response in these patients.
Whitfield ML, Finlay DR, Murray JI, Troyanskaya OG, Chi JT, Pergamenschikov A, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A. 2003;100(21):12319–24.
• Assassi S, Swindell WR, Wu M, Tan FD, Khanna D, Furst DE, et al. Dissecting the Heterogeneity of Skin Gene Expression Patterns in Systemic Sclerosis. Arthritis Rheumatol. 2015;67(11):3016–26. doi:10.1002/art.39289. Partial recapitulation of the SSc skin gene expression subsets by a independent group. Importantly, this study shows that the normal-like group likely represents end stage disease.
Bolen CR, Uduman M, Kleinstein SH. Cell subset prediction for blood genomic studies. BMC Bioinformatics. 2011;12(1):258.
• Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–76. doi:10.1038/ng.3259. Report of the GIANT tissue-specific networks used in Taroni, et al. 2016 and Taroni, et al. 2017.
Chikina M, Zaslavsky E, Sealfon SC. CellCODE: a robust latent variable approach to differential expression analysis for heterogeneous cell populations. Bioinformatics. 2015;31(10):1584–91.
Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7(4):287–9.
Nelms BD, Waldron L, Barrera LA, Weflen AW, Goettel JA, Guo G, et al. CellMapper: rapid and accurate inference of gene expression in difficult-to-isolate cell types. Genome Biol. 2016;17(1):201.
Wong AK, Park CY, Greene CS, Bongo LA, Guan Y, Troyanskaya OG. IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2012;40(Web Server issue):W484–90.
Lemaire R, Farina G, Kissin E, Shipley JM, Bona C, Korn JH, et al. Mutant fibrillin 1 from tight skin mice increases extracellular matrix incorporation of microfibril-associated glycoprotein 2 and type I collagen. Arthritis Rheum. 2004;50(3):915–26.
Gerber EE, Gallo EM, Fontana SC, Davis EC, Wigley FM, Huso DL, et al. Integrin-modulating therapy prevents fibrosis and autoimmunity in mouse models of scleroderma. Nature. 2013;503(7474):126–30.
Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68.
Assassi S, Radstake TR, Mayes MD, Martin J. Genetics of scleroderma: implications for personalized medicine? BMC Med. 2013;11:9.
Allanore Y, Simms R, Distler O, Trojanowska M, Pope J, Denton CP, et al. Systemic sclerosis. Nat Rev Dis Primers. 2015;1:15002. doi:10.1038/nrdp.2015.2.
Greene CS, Troyanskaya OG. Chapter 2: data-driven view of disease biology. PLoS Comput Biol. 2012;8(12):e1002816.
Hintze A, Adami C. Evolution of complex modular biological networks. PLoS Comput Biol. 2008;4(2):e23.
Jeong H, Mason SP, Barabási A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–2.
•• Lofgren S, Hinchcliff M, Carns M, Wood T, Aren K, Arroyo E, et al. Integrated, multicohort analysis of systemic sclerosis identifies robust transcriptional signature of disease severity. JCI Insight. 2016;1(21):e89073. Analysis of publicly available data from SSc cohorts to develop a 4S signature of MRSS severity in SSc patients
Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology. 2000;47(2–3):85–118.
•• Taroni JN, Martyanov V, Mahoney JM, Whitfield ML. A functional genomic meta-analysis of clinical trials in systemic sclerosis: towards precision medicine and combination therapy. J Invest Dermatol. 2017;37(5):1033–41. doi:10.1016/j.jid.2016.12.007. First meta-analysis of molecular gene expression data from SSc clinical trials. Study makes the first attempt to predict improvers across treatments.
Martyanov V, Whitfield ML. Molecular stratification and precision medicine in systemic sclerosis from genomic and proteomic data. Curr Opin Rheumatol. 2016;28(1):83–8.
Whitfield ML. Editorial: plasma and B cell gene signatures: quantitative targeting and monitoring of B cell—depleting therapies in autoimmune diseases in the genomic era. Arthritis Rheum. 2014;66(1):10–4.
Lafyatis R, Kissin E, York M, Farina G, Viger K, Fritzler MJ, et al. B cell depletion with rituximab in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2009;60(2):578–83.
Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21.
• Chakravarty EF, Martyanov V, Fiorentino D, Wood TA, Haddon DJ, Jarrell JA, et al. Gene expression changes reflect clinical response in a placebo-controlled randomized trial of abatacept in patients with diffuse cutaneous systemic sclerosis. Arthritis Res Ther. 2015;17:159. Pilot study of abatacept in SSc with molecular data. Suggest patients most likely to improve on abatacept are those with an inflammatory signature.
• Gordon JK, Martyanov V, Magro C, Wildman HF, Wood TA, Huang WT, et al. Nilotinib (Tasigna™) in the treatment of early diffuse systemic sclerosis: an open-label, pilot clinical trial. Arthritis Res Ther. 2015;17:213. Pilot study of nilotinib in SSc. Study finds that patients most likely to improve on nilotinib are the subset of patients that have high TGFβ signaling at baseline.
Farina G, Lafyatis D, Lemaire R, Lafyatis R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2010;62(2):580–8.
• Rice LM, Ziemek J, Stratton EA, McLaughlin SR, Padilla CM, Mathes AL, et al. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 2015;67(11):3004–15. Longitudinal biomarker for MRSS for use in SSc clinical trials.
• Rice LM, Padilla CM, McLaughlin SR, Mathes A, Ziemek J, Goummih S, et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest. 2015;125(7):2795–807. Pilot study of fresolimumab to target TGFβ activation in SSc patients, which included clinical and molecular gene expression data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Jaclyn N. Taroni declares that she has no conflict of interest. J. Matthew Mahoney declares that he has no conflict of interest. Michael L. Whitfield declares that he has no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Scleroderma
Rights and permissions
About this article
Cite this article
Taroni, J.N., Mahoney, J.M. & Whitfield, M.L. The Mechanistic Implications of Gene Expression Studies in SSc: Insights From Systems Biology. Curr Treat Options in Rheum 3, 181–192 (2017). https://doi.org/10.1007/s40674-017-0072-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40674-017-0072-0