Abstract
Purpose of Review
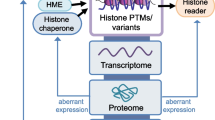
This review will discuss histone modification events that have been recently discovered or significant technical advancements made in the last year. The physiological state of the eukaryotic genome is chromatin. Chromatin is a complex of DNA and primarily histone proteins. The post-translational modification of histones and the occupancy of specific histone variants in a given nucleosome play a central role in the regulation of the genome. Histone variants and modifications exist in complex and unique combinations. The resulting diversity allows for histones to play key roles in almost all cellular processes and their dysregulation is often the root cause of many diseases. In particular, epigenetic alterations have been noted in cancer, are associated with its progression, aggressiveness, and metastasis, and are useful in its prognosis. Thus, a deep understanding of histone variants and modifications is needed to comprehend both normal cellular function and disease.
Recent Findings
Technological advancements have dramatically improved our quantitative understanding of histone modification events, decreased sample requirements, and revealed nuances about how these events work together. Specifically, the miniaturization of ChIP-seq and the continuing use and development of mass spectrometric approaches for histone analysis have revealed interesting and diverse new aspects of chromatin biology.
Summary
This review will explore the causes, mechanisms, and downstream functions of these newly discovered events. We will conclude with an outlook on the future of histone modification analysis and its biological impact.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
• Zhou L, et al. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat Commun. 2016;7:10589. H2B-Ub physically places hDot1L’s active site next to H3K79, allowing it to methylate
•• Zhang B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553–7. One of two publications that independently developed a miniaturized ChIP-Seq method enabling the study of challenging materials. Additionally denoted the unusual state of H3K4me3 in development and its potential as an early zygotic regulator
•• Dahl JA, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–52. One of two publications that independently developed a miniaturized ChIP-Seq method enabling the study of challenging materials. Additionally denoted the unusual state of H3K4me3 in development and its potential as an early zygotic regulator
•• Xiong X, et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat Chem Biol. 2016;12:1111–8. One of several papers from Yingming Zhao’s group which deal with specific acylations. Here, they determined that reading of crotonylation by MOZ and DPF2 is specific over other acylations and further solidifies crotonylation as a unique modification
•• Li Y, et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol Cell. 2016;62:181–93. Further gives context and function to crotonylation and a different method of recognition by AF9 YEATS which also has a preference for crotonylation over other acylations
• Goudarzi A, et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active Gene promoters. Mol Cell. 2016;62:169–80. Both butyrylation and acetylation are present in active promoters, and both act as direct transcription activators. However, butyrylation competes with acetylation and can prevent the binding of specific bromodomain proteins. This paper shows a novel and specific function for butyrylation in regulation and development
• Xie Z, et al. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol Cell. 2016;62:194–206. Histone Lysine b-Hydroxybutyrylation is enriched in active promoters and is linked to the metabolic state of the cell
• Dell’Orso S, et al. The histone variant MacroH2A1.2 is necessary for the activation of muscle enhancers and recruitment of the transcription factor Pbx1. Cell Rep. 2016;14:1156–68. This study shows that macroH2A1.2 is found at muscle-specific enhancers and is required for H3K27 acetylation and Pbx1 recruitment
Pazienza V, et al. Histone macroH2A1.2 promotes metabolic health and leanness by inhibiting adipogenesis. Epigenetics Chromatin. 2016;9:45.
Borghesan M, et al. DNA hypomethylation and histone variant macroH2A1 synergistically attenuate chemotherapy-induced senescence to promote hepatocellular carcinoma progression. Cancer Res. 2016;76:594–606.
Park S-J, et al. MacroH2A1 downregulation enhances the stem-like properties of bladder cancer cells by transactivation of Lin28B. Oncogene. 2016;35:1292–301.
• Latrick CM, et al. Molecular basis and specificity of H2A.Z-H2B recognition and deposition by the histone chaperone YL1. Nat. Struct. Mol. Biol. 2016;23:309–16. YL1 is a specific H2A.Z-deposition chaperone. In this study, they identify the specific residues and molecular interactions required for YL1 recognition and interaction
Jiang P, Huang P, Yen S-H, Zubair AC, Dickson DW. Genetic modification of H2AX renders mesenchymal stromal cell–derived dopamine neurons more resistant to DNA damage and subsequent apoptosis. Cytotherapy. 2016;18:1483–92.
• Weyemi U, et al. The histone variant H2A.X is a regulator of the epithelial-mesenchymal transition. Nat Commun. 2016;7:10711. H2A.X removal activates EMT transcription factors and promotes mesenchymal characteristics. Re-expression of H2A.X partially reverses EMT but increases metastasis
Shimada M, et al. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat Commun. 2016;7:12059.
Segala G, Bennesch MA, Pandey DP, Hulo N, Picard D. Monoubiquitination of histone H2B blocks eviction of histone variant H2A.Z from inducible enhancers. Mol Cell. 2016;64:334–46.
•• Dang X, et al. Label-free relative quantitation of isobaric and isomeric human histone H2A and H2B variants by Fourier transform ion cyclotron resonance top-down MS/MS. J Proteome Res. 2016;15:3196–203. Established a new technique that can distinguish both histone variants and post-translational modifications. Important in that it can quantify different H2A and H2B variants that are difficult to study
• Chen Y, et al. Quantitative mass spectrometry reveals that intact histone H1 phosphorylations are variant specific and exhibit single molecule hierarchical dependence. Mol Cell Proteomics MCP. 2016;15:818–33. H1 phosphorylation occurs in a hierarchical fashion where S172 is first phosphorylated followed by S187, T18, T146, and T154. Phosphorylations of H1.2 (S172 & S187 only) and H1.4 (all) increase during M phase potentially connecting cell cycle to these modifications
•• Zee BM, Alekseyenko AA, McElroy KA, Kuroda MI. Streamlined discovery of cross-linked chromatin complexes and associated histone modifications by mass spectrometry. Proc Natl Acad Sci. 2016;113:1784–9. Expanded upon their previous method for studying histone interacting proteins. This framework and mentality we believe will greatly help future biological research
Gonzales-Cope M, Sidoli S, Bhanu NV, Won K-J, Garcia BA. Histone H4 acetylation and the epigenetic reader Brd4 are critical regulators of pluripotency in embryonic stem cells. BMC Genomics. 2016;17:95.
Zee BM, Dibona AB, Alekseyenko AA, French CA, Kuroda MI. The Oncoprotein BRD4-NUT generates aberrant histone modification patterns. PLoS One. 2016;11:e0163820.
Bhanu NV, Sidoli S, Garcia BA. Histone modification profiling reveals differential signatures associated with human embryonic stem cell self-renewal and differentiation. Proteomics. 2016;16:448–58.
Luense LJ, et al. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin. 2016;9:24.
Steven ZJ, et al. Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol Cell. 2016;62-2:347–61.
Acknowledgements
This research was supported by a grant from the National Insitute of Health, NIH P01 GM085354.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Matthew V. Holt, Tao Wang, and Nicolas L. Young each declare that they have no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Epigenetics
Rights and permissions
About this article
Cite this article
Holt, M.V., Wang, T. & Young, N.L. Recent Advances in Understanding Histone Modification Events. Curr Mol Bio Rep 3, 11–17 (2017). https://doi.org/10.1007/s40610-017-0050-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40610-017-0050-1