Abstract
Large amounts of autism spectrum disorder (ASD) data is created through hospitals, therapy centers, and mobile applications; however, much of this rich data does not have pre-existing classes or labels. Large amounts of data—both genetic and behavioral—that are collected as part of scientific studies or a part of treatment can provide a deeper, more nuanced insight into both diagnosis and treatment of ASD. This paper reviews 43 papers using unsupervised machine learning in ASD, including k-means clustering, hierarchical clustering, model-based clustering, and self-organizing maps. The aim of this review is to provide a survey of the current uses of unsupervised machine learning in ASD research and provide insight into the types of questions being answered with these methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Over the last 75 years, the definition and categorization of autism have varied and changed since psychiatrist Leo Kanner’s first clinical description of autism in 1943. In 2013, the latest revision of the Diagnostic and Statistical Manual of Mental Disorders (the fifth edition, or DSM-5) came out with a revision on the diagnostic criteria for autism. While over the years the definition of autism has expanded to include Asperger syndrome and Rett’s syndrome, this new revision now defines autism explicitly as a spectrum (American Psychiatric Association, 2013). As the definition of autism or now autism spectrum disorder (ASD) changes, our understanding of ASD must evolve and grow as well. With the expansion of autism into a spectrum, we need to better understand where individuals fall within that spectrum. Defining these phenotypes in either behavioral, genetic, or other contexts will begin to allow us to do that. While the expansion of what is considered to be an ASD diagnosis has expanded over the years, this nonetheless discounts the need to find answers and give hope to families and children who live with ASD.
It is estimated that ASD currently affects 1 in 59 children (with males being approximately four times more likely to be diagnosed than females) across the USA according to a surveillance study conducted by the Center for Disease Control and Prevention (CDC). The CDC released these latest numbers on March 31, 2016 (Baio et al., 2018). With the prevalence of ASD increasing anywhere from 6 to 15% each year from 2002 to 2010, it is imperative to understand underlying differences among individuals on the spectrum [REF]. This alone highlights the urgency for comprehensively understanding ASD further.
While there have been many applications of supervised machine learning with ASD data, unsupervised methods have also been used in ASD research. Unsupervised machine learning includes methods such as clustering and dimensionality reduction that aim to find latent structure in data without the help of truth data such as a value or category to be predicted. While some of the methods covered may also be familiar statistical analyses (such as principal component analysis), it is this common structure that ties unsupervised machine learning methods together. Unsupervised machine learning allows researchers to make use of unlabeled data that may not have pre-existing groups or categories. The purpose of the current review is to summarize the use of unsupervised machine learning in the past 21 years of ASD research.
Methods
Endnote and Google Scholar were searched for peer-reviewed articles. Combinations of the search terms “autism,” “ASD,” “autism spectrum disorder,” “unsupervised machine learning,” “clustering,” “phenotypes,” “subgroups,” and “machine learning” were used.
The abstracts and methods sections of the articles were reviewed and an article was included if the following criteria were met: (a) was published in a peer-reviewed journal; (b) included a group of individuals with ASD, autistic disorder, Asperger syndrome, or pervasive developmental disorder (PDD-NOS); and (c) and unsupervised machine learning model was utilized.
While these procedures may not have produced an exhaustive review, findings should be representative of the current trends of unsupervised machine learning in the field of ASD.
Results
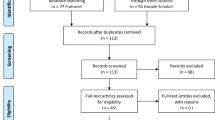
The literature search produced 105 relevant publications. After abstracts and methods were reviewed, 43 research articles were included. See Fig. 1. for PRISMA statement in the Appendix.
Unsupervised machine learning is capable of detecting latent groups or representations in a feature space. Casually referred to as clustering, unsupervised learning partitions data points into groups without having to rely on the label or truth data that is a required input to classification algorithms. While there are many algorithms available for clustering, at their core most clustering algorithms operate by minimizing the distance between cluster members, or, in the case of statistical methods, maximize the likelihood that data points belong to a particular cluster assuming the clusters are described by a probability density function. Perhaps most importantly, the algorithms themselves make no attempt to ascribe meaning to the clusters; that is, left to human analysts. Instead, algorithms simply report the most likely groups explained by the data. The meaning of those groups must be determined by domain experts.
In the cases where a number of clustering techniques were used, papers were categorized by the driving technique deployed. Each section begins with a brief overview of the technique, followed by the reviewed papers pertinent to that section. Due to the vast amount of papers using hierarchical clustering, that specific section is broken down further into studies focused on behavioral outcomes and genetic outcomes. In this paper, many types of data were used including behavioral data, psychological status, developmental factors, and brain imaging. Different types of data may shed light on different latent structures within ASD data, all of which can provide both separate and complementary information and clinical benefit. The various results presented in this paper will hopefully be replicated, confirmed, and compared by experts in the field in order to asses which factors and results have similar structures, and which provide different ways of looking at patterns within ASD data.
Results
K-means Clustering
K-means is a fairly simple technique algorithmically. To start, we initialize k centroids (one for each cluster) to k random data points. From here, each data point within the dataset is assigned to the closest centroid. This assignment to a specific k can be done based on Euclidean distance or any other appropriate distance metric. Once each data point has been assigned, we move each centroid to the average location of the assigned data points. The prior two steps are repeated until cluster membership does not change (MacQueen, 1967). While k-means provides an algorithmically simple way for identifying grouping, the substantial challenge is determining the number of clusters, k, to be modeled. In the absence of a domain expert, several mechanisms exist for statistically determining the most likely k. Popular metrics include the Bayesian information criterion (BIC) (Neath & Cavanaugh, 2012) and the Akaike information criterion (AIC) (Bozdogan, 1987).
In 2016, Lingren and colleagues used data from subjects with ASD and other comorbid disorders (n = 20,658) to cluster subjects into 4–5 clusters (Lingren et al., 2016). Clustering was done using standardized PheWAS codes that distinguish between different symptoms (http://searchhealthit.techtarget.com/definition/ICD-9-CM). A k-means clustering algorithm was used to cluster subjects from each of the three included sites separately. Each site had 3–4 small clusters and one large cluster, and clusters revealed similar comorbidities (represented by similar PheWAS) between site clusters. Cluster 1 included psychiatric problems such as anxiety, hyperkinetic syndrome, OCD, and depression. Cluster 2 included developmental disorders such as dyslexia, lack of coordination, and ear, skin, and other bodily disorders. Cluster 3 was associated with epilepsy and recurrent seizure. The last (and largest) cluster was not defined by any high occurrence of any comorbidities.
Similarly, Hu and Steinberg explore distinguishing clustering phenotypes based on 123 item scores from the ADI-R which also assesses behavioral symptoms of ASD (n = 1,954; Hu & Steinberg, 2009). Since ASD is a spectrum, it covers a wide range of symptoms, both behavioral and physical. Their aim was to reduce the phenotypic heterogeneity of autism by identifying more homogeneous subgroups for gene expression analysis. Hu and Steinberg used a combination of principal component analysis, hierarchical clustering, and k-means clustering to identify subgroups. Furthermore, to estimate the optimal number of clusters, a fitness of merit analysis was completed. Through these techniques, they conclude the number of viable clusters to be four. The four distinguishing groups are those with severe language deficits, those with milder symptoms, those with noticeable savant skills, and those with intermediate severity across all domains. In addition to clustering based on behavioral symptoms, male samples from the first three subgroups were used to explore gene expression analysis.
In another study by Easson et al. (2017), lower triangles from static and dynamic matrices of resting-state fMRIs from 273 males (163 with an ASD diagnosis) from the Preprocessed Connectomes Project (from the ABIDE data set) were clustered using the k-means algorithm. Values for k ranged from 2 to 8. The optimal k was determined to be 2 and consisted of one cluster of 92 participants, and one of 71. Subtype 1 (n = 92) is characterized by stronger static FC between networks and weaker static FC within networks. Weaker stability of dynamic FC was also observed in subtype 1.
Obara et al. use AP clustering on 17 participants (13 males, 4 females) showed 5 well-classified groups, with cluster 1 consisting of potential vitamin B6-responsive subjects with ASD (Obara et al., 2018). The other clusters showed low responses to vitamin B6. k-medoids also showed good classifications with 5 clusters which had nearly identical results to the AP clustering. This classification allows for better classification of people with ASD who will respond to vitamin B6.
Recently, Silleresi et al. (2020) collected measures of language and intellectual ability for 51 verbal children with ASD (ages 6–12; Silleresi et al., 2020). Principal components analysis (PCA) was conducted on all features and two principal components were retained. Using k-means clustering, the data were clustered into 5 distinct clusters. The clusters included all possible combinations of language impairment/disordered intellectual development included in the ICD-11 codes. Three of the clusters consisted of children with normal language abilities, and two consisted of children with language impairment.
Easson et al. found two distinct clusters of functional connectivity (FC) using k-means on neuroimaging data from 145 participants with ASD and 121 typically developing participants (Easson et al., 2019). Subtype/cluster 1 (85 ASD, 54 TD) was defined by stronger FC between networks, especially the default mode network and other networks, and weaker FC within networks relative to subtype cluster 2. This FC information appears to be unique between the subtypes since it was not associated with significantly different IQ, SRS, or ADOS scores between the clusters.
Recently, Hyde et al. examined an online survey looking at the practices and policies with regards to workers with ASD of 285 employers (Hyde et al., 2019). Of the 285 employers surveyed, 58% (166) indicated that they had hired at least one individual with ASD in the past 5 years. K = 5 clusters were fit from the data. All 5 clusters varied with regards to the ASD hiring rate, with cluster 5 having the highest rate for the past 5 years (86%). Clusters revealed that most employers were in the two most extreme clusters (clusters 3 and 5) which represented the least and most favorable policies for ASD related hires, respectively. This indicates that employers may often tend towards one extreme or the other in terms of hiring individuals with ASD. Clusters with high ASD employment rates also tended to have higher rates of employers requiring college degrees for entry-level jobs. Similarly, clusters with the lowest ASD employment rates also had the lowest rates of employers requiring college degrees for entry-level jobs.
Stevens et al. used k-means to identify patterns of challenging behaviors in a sample of 2116 children with ASD (Stevens et al., 2017). They identified 7 clusters to exist, and while the presence of multiple challenging behaviors is common, in most cases a dominant behavior emerged. Furthermore, the trend was also observed when cluster models were trained on the male and female samples separately. The k-means algorithm has shown the potential to create clinically valid clusters for subjects with ASD.
Hierarchical Clustering
Unlike k-means, which learns k distinct clusters without attempting to capture the relationship between them, hierarchical clustering aims to create clusters such that they fall into a network hierarchy. Two approaches exist to do this, the agglomerative approach and the divisive approach. The agglomerative approach or commonly known as the “bottom-up” approach initially treats each datapoint as its own cluster and then combines clusters together until each cluster created falls within an overarching single cluster. Conversely, the divisive approach or “top-down” technique deploys the opposite. To begin, the entire dataset is treated as one large cluster and then splits are made recursively. While the agglomerative approach is more common, both techniques employ a distance metric, similar to k-means, to decide cluster membership and specific linkage criteria are used to determine dissimilarity between clusters.
Behavioral
Sacco and colleagues used questions from 36 developmental, clinical, and family history variables, as well as physical measurements and autistic behaviors, adaptive functioning, and IQ from 245 subjects with ASD (Sacco et al., 2012). Using principal components of the previously mentioned features, hierarchical clustering was used on the factor scores to distinguish clusters. Then, the number of clusters found using agglomerative hierarchical clustering was used as the k in a k-means clustering algorithm. Four clusters were found: Immune Circadian and Sensory which had issues with immune deficiency and circadian and sensory issues (ICS; n = 43); Circadian and Sensory which had higher factor scores for circadian and sensory dysfunction, some neurodevelopmental delays and stereotypic behaviors (CS; n = 44); Stereotypic Behaviors which had verbal and motor stereotypes (S; n = 75); and Mixed which had a mixed pattern and higher neurodevelopmental dysfunction (M; n = 83).
In 1995, Sevin and colleagues used data from 34 children with a diagnosis of PDD (27 of whom met criteria for ASD) on the childhood autism rating scale (CARS), the Ritvo-Freeman real-life rating scale (RLRS), the autism behavior checklist (ABC), the Vineland adaptive behavior scale (Sevin et al., 1995). These variables were used to cluster subjects using Euclidean distance and Ward’s minimum variance method to minimize within-cluster variance. Four clusters were found that best maximizes between-group distance while minimizing within-group distance. Cluster 1 had the highest functioning subjects and most well-developed language skills. Cluster 4 had the lowest functioning with severe social and language impairment as well as social withdrawal. Clusters 2 and 3 were generally between clusters 1 and 4 with cluster 2 showing moderate social and language impairments, mild stereotypes and sensory abnormality, and less evident ritualistic behavior. Cluster 3 had moderate social and language impairments, severe sensory abnormalities and stereotypies, and similar patterns of interaction to cluster 2. Clusters were validated using ANOVAs on measures not included in clustering (age, AAMD, ABS/factor). Tukey’s HSD post hoc tests were also run, showing results in the expected directions.
Working with data from 170 toddlers with ASD between the ages of 18 and 33 months, Ben-Sasson et al. identified sensory subgroups and looked at the difference in affective symptoms across the identified clusters (Ben-Sasson et al., 2008). Five measures were taken on each child, infant/toddler sensory profile (ITSP), the infant-toddler social and emotional assessment (ITSEA), autism diagnostic interview-revised (ADI-R), autism diagnostic observation schedule-generic (ADOS-G), and the Mullen scales of early learning (MSEL). Both scores for the ITSP and ITSEA were created excluding ASD-specific items. These scores were used to perform agglomerative hierarchical clustering analysis. Through this analysis, three subgroups were identified, each having a unique sensory profile. Those in the first cluster display a low frequency of sensory behaviors relative to the other clusters, the second cluster that shows a mixed profile of high frequencies of both under- and over-responsivity but relatively low seeking, and the third group displays the high frequency of all sensory behaviors. A k-means clustering technique was applied to verify the stability of the cluster memberships found using the hierarchical clustering technique. Setting k=3, they found that 84% of the data points kept their cluster membership in the k-means approach. Beyond the identification of sensory profiles within the subgroups, the three clusters differed in their ADI-R scores and gender distribution the most over the other scores or age. Affective symptoms, such as depression/withdrawal, negative emotionality, separation distress, inhibition to novelty, and anxiety, were then examined across clusters. The authors concluded that toddlers with ASD needed to have a sensory assessment completed to evaluate patterns and monitor any affective characteristics. They believe that toddlers who fall into clusters 2 and 3 may better benefit from a sensory-based intervention. And in any case, that clinicians collaborate when it comes to intervention plans as sensory and affective symptoms may be associated, highlighting the value of expert clinical opinions in assessing the application of clusters.
Tadevosyan-Leyfer el al. also used the ADI and ADI-R scores to create a 98-dimensional feature space (Tadevosyan-Leyfer et al., 2003). This space is then reduced via principal components analysis, and subjected to hierarchical clustering as provided by the VARCLUS package in SAS. The resulting model identified 6 latent groups defined by features from domains such as language, sensory, social, and developmental milestones. Given the original feature space, however, these clusters most likely relate to highly correlated groups of questions on the ADI-R, rather than actual subtypes of autism. Nevertheless, this work represents an interesting application of unsupervised machine learning to a standard diagnostic instrument that has typically been used for supervised machine learning.
Similarly, Constantino et al. explored the latent factor structure of ADI-R and SRS data to identify phenotypes in a sample of 226 ASD patients in order to characterize specific domains related to PDD-NOS (Constantino et al., 2004). Factors that drive phenotype formation were identified through standard principal component analysis combined with varimax rotation, which eases interpretation by describing clusters with a linear combination only a few key factors. The simplified factor model then formed the basis of a cluster analysis, which was achieved via the VARCLUS implementation from SAS. Findings confirmed the presence of clusters across social behaviors, repetitive behaviors, and language development, further strengthening the argument that unsupervised machine learning can be used to identify useful subdomains of ASD open in which tailored interventions can be based.
Recently, the DSM changed the diagnosis of autism and related disorders to be classified together on the autism spectrum. The aim of the work conducted by Bitsika et al. is to extend previous research of subtyping ASD with the use of further data sources (Bitsika et al., 2008). Data from 53 preschool and school-age children that were collected over a 4-year span was used in this study. Metrics for this study included the Wechsler scale for full IQ and the VIQ scores, the Vineland adaptive behavior scale (VABS) under certain domains, the Childhood autism rating scale-revised (CARS), and the autism spectrum disorder checklist (ASDBC). Using a hierarchical agglomerative clustering approach, three clusters were defined. Of the three clusters defined, one represents a low-functioning group, one a moderately functioning group, and the last representing a high-functioning group. The authors concluded that this study supports the existence of a spectrum for autism.
A longitudinal study was conducted by Stevens and colleagues on 138 school-age children (Stevens et al., 2000). The aim of this study was to explore if significant subgroups exist, if those subgroups were the same as those discovered in a previous study (Fein et al. 1999), on the same group of children but at preschool age, and which characteristics at preschool age predicted school-age functioning. The inclusion criteria for this study were quite vast and metrics included the wing autistic disorder interview checklist (WADIC)—A, B, and C, the Peabody picture vocabulary test (PPVT), nonverbal IQ ratio, and the Vineland adaptive behavior scales communication and socialization domains. Using a hierarchical agglomerative approach, two subgroups were found. These findings validate those found in their previous study with preschool-aged children. In both studies, groups were partitioned into those that are classified as low functioning and those that are classified as high functioning.
Autism spectrum disorder is often accompanied by physiological symptoms. Doshi-Velez et al. look into the co-occurrence of medical comorbidities in ASD (Doshi-Velez, 2014). This study is comprised of 4927 individuals identified to have at least one International Classification of Diseases, Ninth Revision (ICD-9) code. The scope of these codes was narrowed down to include 45 common categories of the ICD-9 codes. Initially, a time series analysis of each patient’s electronic health records was conducted. This time series was constructed using 30 6-month increments from birth to age 15. Using these two sets of information, a 1350 (30 × 45) dimensional vector was created for each patient. A hierarchical clustering analysis was then performed to identify the existence of clusters. In total, four subgroups were found, 3 small high-morbid subgroups and one that was unable to be further resolved. Of these three high-morbid subgroups, 1 is characterized by seizures, 1 is characterized by psychiatric disorders, and 1 is characterized by more complex system disorders including gastrointestinal disorders and auditory disorders, and infections.
Zheng et al, used measures that capture children’s development (such as cognitive and language abilities) from 188 preschoolers with ASD (Zheng et al., 2020). First, principal components analysis (PCA) was conducted on the measures and nine principal components (PC; which represented over 85% of the variance in the data) were retained. Then, hierarchical agglomerative clustering was performed on the nine PCs. Three clusters emerged from this analysis: cluster 1 had relatively high cognitive and adaptive abilities, and lower severity for social behavior, repetitive behavior, and sensory issues (high functioning). Cluster 2 had similarly high cognitive and adaptive abilities but also had higher severity for social behavior, repetitive behavior, and sensory issues. Cluster 3 had the lowest cognitive, language, and adaptive abilities and had high severity for social behavior, repetitive behavior, and sensory issues.
In addition to behavioral data, auditory comprehension measures can also be used to meaningfully cluster ASD subjects. Rapin and colleagues used test scores on expressive phonology and comprehension of words and sentences were used to cluster 62 7–9-year-old children with ASD into 4 clusters (Rapin et al., 2009). Clusters were characterized by low phonology and low comprehension (cluster 1), low phonology and average comprehension (cluster 2), average phonology and low comprehension (cluster 3), and average phonology and average comprehension (cluster 4), showing that all combinations of phonology and comprehension exist independently.
Behavioral ASD data can also be combined with biomedical and genetic data. Sixty-four autistic children who met the International Classification of Diseases, 10th Edition criteria for PDDs and secondarily received a confirmed diagnosis by the ADI-R were included in a study conducted by Hrdlicka and colleagues (Hrdlicka et al., 2004). This study uses MRI data, as well as, EEG results, and IQ testing, among other metrics, to perform a multidisciplinary approach to subtyping. Within the MRI scans, measurements focused on cortex thickness, size of the corpus callosum, hippocampus, caput of the caudate nucleus, and amygdala. A hierarchical clustering analysis was performed using the MRI data to discover the existence of four clusters. Within these clusters, further tests were performed (one-way ANOVA, chi-square test.) to establish the soundness between clusters.
In the study by Pichitpunpong et al., a cluster analysis on the ADI-R scores from 85 individuals with ASD revealed four ASD subgroups with various dysregulated genes (identified via transcriptome profiling; Pichitpunpong et al., 2019). DBI is identified as a novel candidate protein for ASD with severe language impairment.
Genetic
Other studies focus mainly on genetic and biomedical data. Obafemi-Ajayi et al. used an agglomerative hierarchical clustering approach to a mixed dataset of both numerical and categorical features of 213 ASD subjects (Obafemi-Ajayi et al., 2015). This dataset is a subset of the Simon simplex collection project, which includes genetic and other biomedical data, and was narrowed down from 35 features to 24 features to produce more homogenous meaningful subgroups, 7 categorical, and 17 numerical. To validate cluster findings, an ensemble cluster validation was conducted to ensure compactness and separateness. Using a majority vote based on outcomes from the ensemble validation, the optimal number of clusters is determined to be two. While the algorithm identifies two main clusters, a third outlier subgroup does exist and appears to be meaningful. They additionally believe that the two predominate subgroups can be divided further to reveal additional subgroups. The aim of this study was to identify homogeneous groups to aid in genetic studies.
Veatch et al. used seven different clustering methods (i.e., k-means, agglomerative hierarchical, model-based, partitioning around medoids, divisive hierarchical, self-organizing tree algorithm, and clustering large applications) were used to create between 2 to 15 clusters (Veatch et al, 2013). Agglomerative hierarchical clustering was selected as the most valid clustering method which created two clusters, one with 818 data points, and the other with 443. The agglomerative coefficient was 0.78 which indicates a strong clustering structure. In comparison between the two clusters, individuals with severe social and communication deficits as well as those who were relatively young at the time of examination were generally placed in the larger cluster, leading to the conclusion that the larger cluster represents more severe cases of ASD. These two clusters were further subgrouped into 10 clusters, with statistically significant differences in head circumference and ADI-R RRB scores across clusters. Results were replicated with a separate data set which revealed two main clusters and 15 sub-clusters, with significant differences between the “more severe” and “less severe” groups in all variables except head circumference.
In Kurochkin et al., 1366 metabolites in the prefrontal cortex gray matter of 32 individuals with ASD and 40 controls were clustered using hierarchical clustering (Kurochkin et al., 2019). Fifteen percent of these metabolites clustered into 16 metabolic pathways, 10 of which were altered in the urine and blood of ASD individuals, indicating that new diagnostic tests focusing on these metabolites could be developed.
Smith et al. performed hierarchical clustering on the concentration of various metabolites associated with ASD (from the Children’s Autism Metabolome Project; Smith et al., 2019). Metabolites were log base 2 transformed and z-scored prior to analysis. A dissimilarity measure of 1-abs(Pearson correlation) was used to calculate distances for clustering. Three clusters of metabolites emerged. Cluster 1 was the glycine cluster, cluster 2 was the BCAA’s and phenylalanine. Cluster 3 contained glutamate and aspartate. Differences in the metabolites of children with ASD and typically developing children could leave to actionable metabolic tests that support early diagnosis of ASD.
In Duffy and Als, hierarchical and k-means clustering using the NbClust package in R was performed on EEG data from 430 ASD individuals (Duffy & Als, 2019). Forty previously defined features (EEG coherence factors that reliably separated ASD from neurotypical controls as well as subjects with Aspergers from other subjects with ASD) were used in the clustering analyses. For both the hierarchical and k-means clusters, the optimal number of clusters was two. However, the hierarchical clustering solution was accepted because there was stronger agreement the two was the ideal number of clusters. Twenty-four of the 40 factors were significantly different between clusters. There were no differences between the two clusters on gender or age. One cluster (cluster 2) seems to represent subjects with Asperger-like behavioral characteristics.
Model-based Clustering
Another approach to clustering is that which is model-based. Model-based clustering assumes the underlying data fit a probability distribution or mixture of probability distributions. Each one of these distributions is then said to represent a specific cluster within the data. Gaussian mixture models, expectation–maximization (EM), and latent class clustering are all examples of model-based clustering.
Using Gaussian mixture models, Bekele et. al. took data from ten subjects with ASD were asked to identify the emotions of a VR avatar (Bekele et al., 2013). Sixteen extracted features from physiological data collected during the VR session (i.e., EEG, PPG, SKT, and GSR) were mapped using principal component analysis to reduce dimensionality. Using the first two components, k-means and Gaussian mixture models were applied and two clusters of trials (correct and incorrect) were found. Results showed differences in how individuals with ASD processed and discriminated emotions. Pairs of emotions like contempt/disgust, and fear/surprise were confusing for subjects with ASD. However, some instances suggest that subjects with ASD recognized certain expressions with greater accuracy than their control counterparts.
Similarly, Uljarevic et al. look to identify sensory subtypes, as well as, investigate the relationship between identified subtypes and anxiety levels (Uljarevic et al., 2016). This small study was conducted with information from 57 children aged 11–17 years old with ASD. Using model-based cluster analysis, they identified three sensory clusters, sensory adaptive, sensory moderate, and sensory severe. Those who were classified as sensory moderate or severe had significantly higher anxiety scores than those who are sensory adaptive. These results agree with other clustering studies that support the spectral nature of ASD diagnoses.
Lane et al. leveraged model-based cluster analysis combined with Bayesian information criteria to also identify sensory phenotypes based on the short sensory profile (Lane et al., 2010, 2014). The 2010 study among 54 children found the existence of 3 sensory subtypes grounded in taste, smell, and movement, which had a correlation to maladaptive behavior and general communication ability. The 2014 study augmented this work to analyze clustering behavior among 228 children between the ages of 2–10 years with ASD. The short sensory profile provided the feature space for the clusters, which were then compared to characteristics such as IQ. Using this approach statistically meaningful subtypes was identified corresponding to different sensory issues, as well as different severities for specific issues. A conclusion of this work is that clustering techniques provide a meaningful direction for tailored intervention, but should be expanded to include features from a wider variety of cognitive and physiological domains. This work represents an extension of the work by the authors in (Lane 2011), which laid the foundation for sensory-based clustering, though on a much smaller sample size of 30 children.
Model-based clustering was also used to explore issues with physiological comorbidities, in this case, epilepsy. Conservative estimates suggest that epilepsy occurs in ~25% of those with ASD. Due to this, Cuccaro et al. looked to explore the relationship between ASD and epilepsy (Cuccaro et al, 2012). Three different latent class cluster analyses were conducted to identify subgroups. Results from the ADI-R, the Vineland adaptive behavior scales, and the aberrant behavior checklist were used as a measure in this study. The first analysis was done on the entire sample size of 577 individuals age 4–21 with ASD, 64 of these individuals also have co-occurring epilepsy. Two additional analyses were conducted on subsets of this data. The first analysis of the overall dataset returned a five cluster model. Cluster 1 identified those who are globally impaired, cluster 2 represented those with early-onset of behaviors, cluster 3 those with language regression, and high-functioning individuals are represented in clusters 4 and 5. Cluster 2 had the highest prevalence of epilepsy at 29% and clusters 4 and 5 had the lowest prevalence with 8%. The second analysis was conducted on an age-restricted subset of only those aged 10–21. This dataset included 281 individuals. The purpose of this restriction was due to the fact that epilepsy usually peaks at one of two specific times, either early onset, before age 5, or late-onset, in adolescence. Also using latent class cluster analysis, four clusters were detected. As in the first findings, these clusters differentiated in severity and mirrored those found in the overall dataset. The final analysis was conducted only on those 64 individuals with co-occurring epilepsy. This analysis found a three cluster model. Cluster 1 was those with language regression, cluster 2 was those classified as globally impaired, and cluster 3 represented those that are high functioning. For both, the overall and age-restricted LCCA analyses, clusters defined by the early age of recognition and high rates of cases with repetitive objects use and unusual sensory interests contained the highest frequency of cases positive for epilepsy. Due to the frequency of these features, it’s proposed that this combination leads to a particular class where increased seizures/epilepsy are more likely. These features and behaviors are also notably common to ASD but may be part of the epilepsy phenotype
Imaging data can be used with model-based clustering as well. An et al. used synthetic fMRI data was simulated to demonstrate the ability of a novel multi-view EM method that incorporates co-training into the EM framework (An et al., 2010). This method analyzes both white and gray matter (from fMRI and DTI, which are assumed to be independent) in the brain to determine subnetworks of connectivity in subjects with ASD. This type of EM allows tw EM procedures to happen simultaneously. For their experimental analysis, subjects were shown points tracing out the scrambled human motion (which looks like non-biological motion). Restricting data to the right hemisphere, the mv-EM classification was run 10 times for each subject. The initial parameters for the E step were based on a random subnetwork labeling of the data. Clustering results were similar to their hypothesized pathways with subnetwork 1 comprises the superior temporal sulcus (STS), inferior frontal gyrus (IFG), and extrastriatal body area (EBA), subnetwork 2 comprises the amygdala (AMY) and fusiform gyrus (FFG), and subnetwork 3 comprises the STS and the medial prefrontal cortex (mPFC).
In 2018, Zheng, Grove, and Eapen used latent profile analysis to cluster data from 2759 participants from the Simons Simplex Collection (2384 males; Zheng, Grove and Eapen, 2019). Clustering was performed on 43 items from the Revised Repetitive Behavior Scale (RBS-R). Using various measures of model fit (such as BIC, AIC…) the three cluster solution was determined to be optimal. These three groups represented low, medium, and high severity groups. The three groups differed not only on the clustered items, but also on similar measures such as the ADI-R RRBI, and the ADOS RRBI. The authors state that this result helps in the detection of subtypes in ASD.
In 2019, 34 items from the short sensory profile 2 (SSP-2) were collected for 248 students from the longitudinal study of Australian students with autism (LASA; Simpson et al., 2019). Normal mixture models were fit using Dirichlet process mixture models since the data had both continuous and ordinal categorical data. Two major clusters were found. One cluster, called “uniformly elevated” had 182 participants with elevated differences across all domains in the SSP-2. The other cluster was smaller (n = 66) who had typical scores for seeking and registration, but elevated scores in sensitivity and avoiding. These two distinct clusters are helpful because, as the authors point out, the heterogeneity of symptoms among children on the spectrum can make individualized support difficult. Establishing subtypes through clustering allows for more tailored treatments based on symptoms.
In Stevens et al. (2019), a sample of children with ASD (N = 2400) was clustered first using Gaussian mixture modeling. This method revealed 16 subgroups. These subgroups were further combined using hierarchical agglomerative clustering which suggested that there are two overlying behavioral phenotypes with unique deficit profiles. Each of these two groups is made up of subgroups that have different levels of deficit severity. Regression analyses on this dataset revealed notably high R2 values for a model relating ABA treatment hours (intensity) and the number of mastered learning objectives (performance). These values are much higher than in typical datasets.
Model-based clustering allows for more accurate cluster recognition when model assumptions are relatively accurate. When researchers have expertise or data that suggests that cluster shapes have a particular distribution, model-based clustering, unlike k-means, allows for the inclusion of that information. However, when model assumptions are inaccurate, it is possible that model-based techniques will not be robust to the violation of those assumptions. Therefore, it is important for future work to include empirical and/or expert-based information about model assumptions.
Self-organizing Maps
Self-organizing maps (SOMs) are a form of artificial neural networks that is capable of taking high-dimensional input data and mapping it to a low-dimensional space. Often the number of dimensions mapped to are small (two or three), to facilitate data visualization and human interpretability. A key property of SOMs is that they maintain spatial relationships among the input data in the transformed output space. Therefore, items that are mathematically “close” to each other in the input feature space remain close to each other in the output space. This is convenient for human interpretability as visualizations to depict similar groupings of data points can easily be generated, and these groupings can be thought of as analogous to clusters learned by other unsupervised learning algorithms. Though SOMs are artificial networks, training differs from classic feed-forward networks as backpropagation is not employed, as this requires labeled data to calculate gradient updates in order to minimize error observed at the output layer of the network. Instead, SOMs learn synaptic weights such that sets of neurons respond more or less strongly to different spatial characteristics of input data, effectively determining group membership for data presented to the network.
In 2011, Wiggens and colleagues did SOM analysis on eyes-open resting-state fMRI data from 80 participants (39 with ASD, 41 without) found that adolescents with ASD have weaker connectivity between the posterior hub of the default network and the right superior frontal gyrus as well as smaller increases of connectivity with age (Wiggins et al., 2011). The default network usually increases in activity when there is no focused task, and decreases in activity when there is a cognitively demanding one. It was also found that subjects with ASD had a different developmental trajectory of the default network. These results held up even when excluding medicated subjects as well as in models where verbal and nonverbal cognitive functioning was used as a covariate.
Similarly, in Tovar et al., 27 children with ASD and 34 typically developing children were asked to examine novel objects that had a given label (such as “Tuka” or “Rako”) and then one by one, they were asked to label other similar or dissimilar objects (Tovar et al., 2020). A self-organizing map (SOM) was fit to the pattern of responses generated by the children on this task. There were two resulting clusters and three outlier points. The groups generally matched a shape and color subgroup, and a shape bias group. The shape and color cluster generally extended the novel label to items that matched in color or shape with the novel object. The results suggest that children with ASD are more likely to extend novel word labels to objects based on color. This result is confirmed with a simulation study that produced similar results.
Node-based Resilience Clustering
Node-based resilience (NBR) clustering uses a graph-theoretic framework to represent and split data points into groups (Matta et al., 2018). First, graphs representing the data are created by creating a node (or vertex) for each observation. Nodes are connected via edges to their k nearest neighbors, resulting in the final graph. An optimal set, S, of so-called “attack nodes” are then removed which disconnect the graph into multiple, non-connected clusters of nodes. If a specific k is desired, clusters can be merged or separated until the desired k is reached. Nodes that are a part of the attack set can either be excluded from classification or can be assigned to the nearest cluster. Various measures of node-based resilience (such as Vertex attack tolerance, integrity, and tenacity) can also be calculated (see Matta et al., 2018 for an overview of these measures)
In Matta et al., 2018, 2680 subjects (2,316 males) between the ages of 4 and 17 were examined. Features included 36 normalized phenotype features (see Matta et al., 2018 for detailed list) from the Simons Simplex Collection were used for clustering. Additional features were compared between resultant clusters to validate the differences between clusters. One hundred and forty-four different clustering outputs were created based on combinations of the following characteristics: number of clusters (k between 2 and 5), using reduced or full features (33 vs 36), NBR measure (VAT, Integrity, or Tenacity), kNN connectivity of graph (kNN = 2,3,4), attack node assignment to clusters (assigned, or excluded). The top seven clustering outputs were chosen and examined. Four out of the seven outputs had k=5, indicating this configuration to be optimal. In general, there were no statistically significant differences in age or gender between clusters. The percentage of Caucasian subjects did differ significantly. Focusing on one specific output (kNN = 2, integrity, 3 clusters), three clusters emerged that corresponded to low, medium, and high severity of symptoms (based on non-clustered features such as overall ABC score, and ADOS CSS) with the high severity cluster having the lowest number of members (7%), the medium severity cluster had 22% of the original sample, and the low severity cluster had the most members with 71% of the original sample. The results from another output (kNN = 2, tenacity, 5 clusters) also showed large effect sizes for differences between clusters on non-clustered features.
Autoencoders
Autoencoders are a type of unsupervised (or semi-supervised) neural network that consists of two parts: the encoder and the decoder. The encoder takes data and creates a latent representation of the data; the decoder takes that latent representation and tries to recreate the original output. In general, the loss of the model is calculated using the reconstruction error. The latent representations created by the encoder are analogous to nonlinear principal components analysis (or regular PCA if the activation functions are purely linear).
Jaiswal et al. used structural brain MRIs (sBMRI) from nine different sources (12,387 images, 1,127,217 images after applying rotations and transformations)—including the ABIDE-I data set—to train a deep convolutional autoencoder (Jaiswal et al., 2018). Convolutional autoencoders (CAEs) have a similar structure to autoencoders, but the encoder performs convolution and the decoder performs deconvolution. Three CAEs were fit to create latent representations of the sBMRIs, which were then used in supervised classifiers to classify Healthy Controls vs. subjects with various diagnoses. In the ABIDE-I dataset, classifiers on the latent representations had AUC-ROC scores between 0.57 and 0.66. Constructing the latent representation for the sBMRIs is substantially faster than other popular methods such as FreeSurfer which takes 20-47 hours per image vs about 0.50 seconds for the CAEs.
Similarly, Pinaya et al. built a deep autoencoder (104-100-75-100-104) trained in a semi-supervised manner using data from 1,13 healthy control (HC) subjects (Pinaya et al., 2019). The autoencoder was semi-supervised because the loss function included both the reconstruction loss (unsupervised), as well as the cross-entropy for age and sex prediction (supervised). A deviation metric (the average squared error for all brain regions) was calculated for each subject. This deviation metric was also calculated for 83 subjects with ASD (from the ABIDE dataset; not used in the training of the autoencoder). Researchers reported a significant difference between the deviation metrics of HCs vs subjects with an ASD diagnosis. Significant differences between subjects with ASD and HCs were found in the choroid plexus, cuneus, putamen, and cerebellum cortex. Using an autoencoder trained on HCs allowed for improved detection of ASD diagnosis.
Recently, eye-tracking scan paths from 59 children from a French school were fed into a deep autoencoder in order to reduce the dimensionality of the data (Elbattah et al., 2019). Each 100×100 grayscale image of a scan path was fed into the autoencoder for training over 100 epochs. The dimensionality was reduced from 10,000 to 500 features. The resulting encodings were used for the subsequent k-means (k = 2:4) clustering analysis. K = 2 had the best fit (measured via silhouette scores). Cluster 2 had a notably higher percentage of individuals with ASD.
Autoencoders allow large datasets with many features to be condensed in an efficient manner, the latent representations created can be used for clustering, or for classification with less computational complexity than doing the same tasks with the full dataset.
Other
Within this section fall all reviewed papers that did not fit into any of the above categories.
Ensemble methods combine the predictive power of multiple models in order to make stronger predictions. Shen et. al. analyzed behavioral data from 394 subjects (358 after exclusion) with PDD answered 93 questions (Shen et al., 2007). Questions were grouped into features. These features were used to perform k-means, hierarchical, and EM clustering. The methods all found between 3 and 7 clusters. The 3-cluster solution was the most optimal for fitness and stability in most cases (the exception being that the 5-cluster solution was most stable for hierarchical clustering). A 6×1 vector was used to represent the optimal solution for fitness and stability for each clustering technique for each subject. Four combinations of solutions represented a large majority of subjects. Clusters related to clinical diagnosis with cluster 1 being dominated with autism diagnosis and cluster 2 being dominated by Aspergers. The diagnosis did not differ significantly between clusters 3-and 4.
While the above categories are the most prominently used in unsupervised learning, it is not to say that new techniques could not be developed. Ingalhalikar et al. aim to create a novel unsupervised learning technique using imaging data to apply to two separate clinical populations, ASD, and schizophrenia (Ingalhalikar et al., 2012). Multi-edge graphs are created for each population and then spectral clustering is performed on each of these graphs. The validity of cluster membership is tested using a concept that determines the certainty of a subject’s current assigned cluster membership against the pull from other clusters, this is noted as “holding power.” Strictly looking at the ASD population within this study, 33 individuals with ASD were looked at and 21 age-matched individuals without ASD were used as controls. DWI images (diffusion-weighted magnetic resonance imaging), verbal IQ, social responsiveness scale (SRS), social communication questionnaire (SCQ), clinical evaluation of language fundamentals (CELF), full-scale IQ, ADOS, and perceptual reasoning index (PRI) scores were used as data. Using the abovementioned approach, this novel technique split the data into two clusters with 92% accuracy against the truth data.
Work has also been done to provide a rigorous framework to assist in cluster selection when using one or more clustering algorithms that have tuning parameters. Nguyen et. al. propose an ensemble validation paradigm to assist in finding clinically optimal clusters. This method can be used with many different kinds of clustering algorithms. For each of i clustering outputs that is produced, j internal clustering validation metrics (CVMs) are calculated. Cij refers to the jth CVM for the ith output. These are then ranked, and the top r outputs for each CVM are selected. All outputs except the top r for each CVM are given a rank of 0. The optimal scheme has the highest sum of ranked CVMs. Applied to ASD data from the SSC dataset, 4 optimal schemes were identified, 3 of which were binary clusters with similar labels (Nguyen et al., 2018).
While the prior above categories discussed are the leading techniques used, not all research problems call for the same approach. Due to the nature of the research being inherently exploratory, sometimes a combination of techniques is required to address different aspects of the task at hand, this combination creates an ensemble method. Additionally, as more data is collected and new data explored, new clustering techniques may be needed to suit certain problems better, as demonstrated by Ingalhalikar et al. above.
Discussion
This paper aimed to review the body of scientific literature that applies unsupervised machine learning to ASD research. A total of 43 articles were identified and reviewed. Overwhelmingly, hierarchical clustering was the most widely used technique, followed by model-based techniques, k-means, self-organizing maps, and others. Because ASD is a spectrum, it is imperative to try to distinguish and understand the inherent latent subgroups that do exist. With an unsupervised technique, the goal is to distinguish clusters that are homogeneous on some scale. Within ASD, researchers have looked to identify subgroups in the areas of behavioral, genetic, sensory, and neurological. Hierarchical clustering also offers a practical advantage for clinicians who may need flexibility in choosing the number of groups they want to work with. While hierarchical clustering can provide an optimal number of clusters, the inherent structure of the model allows researchers and clinicians to see and take advantage of the hierarchical relationships between the clusters and combine or split clusters as needed. For example, even if the optimal number of clusters is 5, if clinicians want to create two separate treatment plans, diagnoses, or interventions, they can do so using hierarchical clustering.
The studies included in this review reveal many strengths in the literature. Clustering methods are useful, especially since the change from the DSM-IV to the DSM 5 which classifies autism and other related disorders as a spectrum. While many previous diagnoses, like Asperger’s syndrome, are now under one diagnosis, people with different symptoms and levels of severity may need different types of treatment and support. Classifying patients with respect to both behavioral and biological features will allow clinicians to give more targeted treatment. These groups can also be used to help clinicians understand more about the mechanisms underlying treatment plans by allowing them to see who is benefiting most from different types and intensity of treatment and support. Similarly, grouping patients with ASD may allow researchers to find metrics that improve the accuracy and latency of diagnosis. Because early diagnosis and treatment have a positive impact on individual outcomes, any metric that can help reduce time to diagnosis could have a positive effect. Future research can attempt to replicate and confirm the patterns observed in the studies covered by this review, and can provide more specific insight into possible latent structures that can be leveraged by clinicians in order to provide more targeted treatment.
There are also some areas for future improvement in the field. While many studies agree on the general optimal number of clusters (around 2–4), very little work has been done examining whether cluster membership is consistent across studies, or across different metrics (behavioral vs. biological). This review shows that the repeated occurrence of 2–4 clusters, even on vastly different input features, may suggest a deeper, more consistent structure across multiple data types (such as behavioral, genetic, and physiological data). Such consistency would allow for even greater degrees of personalization of interventions, in addition to the development of predictive and statistical models for informing and predicting treatment and its progress by integrating data from multiple domains. Larger scale studies are also needed in order to examine whether cluster structure remains steady over larger populations of individuals with ASD. Studies included in this review generally had small sample sizes (see Appendix.Table 1). While small studies can lead to useful insights, they also risk overlooking minority groups, underrepresenting heterogeneity in the data, and establishing unstable patterns of clusters that do not apply to a larger population. Similarly, studies that look at data points that are not individuals (e.g. clustering genes, or other biological markers), larger scale studies should be done to see whether the patterns that emerged hold in larger populations. We also recommend further examination of whether changing treatment in response to cluster membership has an impact on patients’ outcomes.
Future studies using unsupervised machine learning in ASD research could look at differential reactions to ABA in general (as well as different ABA components) between and within established clusters. More studies could apply unsupervised methods to biophysiological data (like MRIs, EEGs, biomarkers...) in order to examine whether observed individual behavioral and sensory differences are related to biophysiological differences. So far, few studies look at whether cluster structure is consistent across multiple data domains. More studies could also be done to examine the different sensory triggers that individuals with ASD have (i.e., some like “cold and crunchy” foods, some like only soft food. Some don’t like certain fabrics because they feel uncomfortable on their skin) to see if common co-occurrences/patterns emerge or whether they are associated with behavioral or biophysiological differences between patients.
Methodologically, more density-based clustering methods (such as Gaussian Mixture Models) could be used. Similar to hierarchical clustering, density-based methods often give researchers and clinicians flexibility in the membership of individual data points to different clusters since they often give soft (or probabilistic) cluster assignments. Because flexibility is important in a clinical setting, density-based methods should be further explored. In addition, while clustering is a large part of unsupervised machine learning, it is not the only option. Our review describes promising ways that other unsupervised methods (like autoencoders) can be used to deal with high-dimensional data, which is especially prevalent in biophysiological data. These methods should be further explored so that this field can take advantage of large and high-dimensional data that would otherwise prove unwieldy without these methods.
The large amount of unlabeled data collected from patients with ASD has the potential to provide clinicians, patients, and families with valuable information about managing the symptoms of ASD. Many papers in this review have used small subsections of ASD data to create clusters that can help with treatment and diagnosis. In the future, collaborations or larger data sets could provide more consistent and widely applicable standards for clusters. In addition to systematic reviews such as this one, we recommend that a large-scale meta-analysis of unsupervised machine learning applications to ASD be undertaken to gain additional insight into clusters, and in particular if a stable set of high-level clusters, regardless of the specific feature space, can be identified and leveraged for future analyses. Since ASD is now considered a spectrum disorder, methods like hierarchical clustering can be especially useful when categorizing “subtypes” of ASD because the hierarchical structure allows for subtypes of subtypes which gives insight into the relationship between clusters, as well as provides flexibility for clinical applications. In conclusion, unsupervised machine learning provides a practical way for clinicians and researchers to take advantage of individual differences within people with ASD in order to improve treatment, and better understand individuals at different points in the spectrum. The nature of ASD as a spectrum disorder allows for a diverse population of individuals who share a diagnosis, identifying subtypes in symptoms, behaviors, and biological markers may help both clinicians and individuals make the most of therapeutic interventions, provide more effective support, and help better identify strengths and limitations that individuals with ASD have. Further research is needed in order to consolidate the knowledge generated by the many smaller scale studies reviewed in this paper and provide evidence that patterns established by these studies hold up across different populations. There is a substantial and compelling need for studies with larger ASD datasets, as well as studies that look at whether differential treatment plans for clusters identified in the research have an impact on individual outcomes. Overall, ASD research is uniquely suited for unsupervised methods because of the diversity of individuals who share a diagnosis, and both current and future uses of unsupervised machine learning in ASD research are promising.
References
An, M., Ho, H. P., Staib, L., Pelphrey, K., and Duncan, J. (2010). Multimodal MRI analysis of brain subnetworks in autism using multi-view EM. In 2010 Conference Record of the Forty Fourth Asilomar Conference on Signals, Systems and Computers, pages 786–789. IEEE.
Association, AP et al. (2013). Diagnostic and statistical manual of mental disorders (DSM-5) American Psychiatric Pub.
Baio, J., Wiggins, L., Christensen, D. L., Maenner, M. J., Daniels, J., Warren, Z., Kurzius-Spencer, M., Zahorodny, W., Rosenberg, C. R., White, T., et al. (2018). Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, united states, 2014. MMWR Surveillance Summaries, 67(6), 1.
Bekele, E., Zheng, Z., Swanson, A., Crittendon, J., Warren, Z., & Sarkar, N. (2013). Understanding how adolescents with autism respond to facial expressions in virtual reality environments. IEEE transactions on visualization and computer graphics, 19(4), 711–720.
Ben-Sasson, A., Cermak, S., Orsmond, G., Tager- Flusberg, H., Kadlec, M., & Carter, A. (2008). Sensory clusters of toddlers with autism spectrum disorders: Differences in affective symptoms. Journal of Child Psychology and Psychiatry, 49(8), 817–825.
Bitsika, V., Sharpley, C., & Orapeleng, S. (2008). An exploratory analysis of the use of cognitive, adaptive and behavioural indices for cluster analysis of ASD subgroups. Journal of Intellectual Disability Research, 52(11), 973–985.
Bozdogan, H. (1987). Model selection and Akaike’s information criterion (AIC): The general theory and its analytical extensions. Psy- chometrika, 52(3), 345–370.
Constantino, J. N., Gruber, C. P., Davis, S., Hayes, S., Passanante, N., & Przybeck, T. (2004). The factor structure of autistic traits. Journal of Child Psychology and Psychiatry, 45(4), 719–726.
Cuccaro, M. L., Tuchman, R. F., Hamilton, K. L., Wright, H. H., Abramson, R. K., Haines, J. L., Gilbert, J. R., & Pericak- Vance, M. (2012). Exploring the relationship between autism spectrum disorder and epilepsy using latent class cluster analysis. Journal of autism and developmental disorders, 42(8), 1630–1641.
Doshi-Velez, F., Ge, Y., & Kohane, I. (2014). Co-morbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics, 133(1), e54–e63.
Duffy, F. H., & Als, H. (2019). Autism, spectrum or clusters? an eeg coherence study. BMC neurology, 19(1), 27.
Easson, A. K., Fatima, Z., and McIntosh, A. R. (2017). Defining subtypes of autism spectrum disorder using static and dynamic func- tional connectivity. bioRxiv, page 198093.
Easson, A. K., Fatima, Z., & McIntosh, A. R. (2019). Functional connectivity-based subtypes of individuals with and without autism spectrum disorder. Network Neuroscience, 3(2), 344–362.
Elbattah, M., Carette, R., Dequen, G., Guerin, J.-L., & Cilia, F. (2019). Learning clusters in autism spectrum disorder: Image- based clustering of eye-tracking scanpaths with deep autoencoder. In 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (pp. 1417–1420). IEEE.
Hrdlicka, M., Dudova, I., Beranova, I., Lisy, J., Belsan, T., Neuwirth, J., Komarek, V., Faladova, L., Havlovicova, M., Sedlacek, Z., et al. (2005). Subtypes of autism by cluster analysis based on structural MRI data. European child & adolescent psychiatry, 14(3), 138–144.
Hu, V. W., & Steinberg, M. E. (2009). Novel clustering of items from the autism diagnostic interview-revised to define phenotypes within autism spectrum disorders. Autism Research, 2(2), 67–77.
Hyde, K., Griffiths, A.-J., Giannantonio, C., Hurley-Hanson, A., Mathur, S., and Linstead, E. (2019). Exploring the landscape of employers for individuals with autism spectrum disorder. In 2019 18th IEEE International Conference On Machine Learning And Applications (ICMLA), pages 1961–1967. IEEE.
Ingalhalikar, M., Smith, A. R., Bloy, L., Gur, R., Roberts, T. P., & Verma, R. (2012). Identifying sub-populations via un- supervised cluster analysis on multi-edge similarity graphs. In International Conference on Medical Image Computing and Computer-Assisted Interven- tion (pp. 254–261). Springer.
Jaiswal, A., Guo, D., Raghavendra, C. S., and Thompson, P. (2018). Large-scale unsupervised deep representation learning for brain structure. arXiv preprint arXiv:1805.01049.
Khilnani, D., Piech, C., and Thitte, S. (n.d.) MRI based machine learning for identification of novel subtypes in autism.
Kurochkin, I., Khrameeva, E., Tkachev, A., Stepanova, V., Vanyushkina, A., Stekolshchikova, E., Li, Q., Zubkov, D., Shichkova, P., Halene, T., et al. (2019). Metabolome signature of autism in the human prefrontal cortex. Communications biology, 2(1), 1–10.
Lane, A. E., Molloy, C. A., & Bishop, S. L. (2014). Classification of children with autism spectrum disorder by sensory subtype: A case for sensory-based phenotypes. Autism Research, 7(3), 322–333.
Lane, A. E., Young, R. L., Baker, A. E., & Angley, M. T. (2010). Sensory processing subtypes in autism: Association with adaptive behavior. Journal of autism and developmental disorders, 40(1), 112–122.
Lingren, T., Chen, P., Bochenek, J., Doshi-Velez, F., Manning-Courtney, P., Bickel, J., Welchons, L. W., Reinhold, J., Bing, N., Ni, Y., et al. (2016). Electronic health record based algorithm to identify patients with autism spectrum disorder. PloS one, 11(7), e0159621.
Matta, J., Zhao, J., Ercal, G., & Obafemi-Ajayi, T. (2018). Applications of node-based resilience graph theoretic framework to clustering autism spectrum disorders phenotypes. Applied network science, 3(1), 38.
Neath, A. A., & Cavanaugh, J. E. (2012). The Bayesian information criterion: Background, derivation, and applications. Wi- ley Interdisciplinary Reviews: Computational Statistics, 4(2), 199–203.
Nguyen, T., Nowell, K., Bodner, K. E., and Obafemi- Ajayi, T. (2018). Ensemble validation paradigm for intelligent data analysis in autism spectrum disorders. In 2018 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), pages 1–8. IEEE.
Obafemi-Ajayi, T., Lam, D., Takahashi, T. N., Kanne, S., and Wunsch, D. (2015). Sorting the phenotypic heterogeneity of autism spectrum disorders: A hierarchical clustering model. In 2015 IEEE Conference on Computational Intelligence in Bioinformatics and Computa- tional Biology (CIBCB), pages 1–7. IEEE.
Obara, T., Ishikuro, M., Tamiya, G., Ueki, M., Yamanaka, C., Mizuno, S., Kikuya, M., Metoki, H., Matsubara, H., Nagai, M., et al. (2018). Potential identification of vitamin B6 responsiveness in autism spectrum disorder utilizing phenotype variables and machine learning methods. Scientific reports, 8(1), 14840.
Pichitpunpong, C., Thongkorn, S., Kanlayaprasit, S., Yuwattana, W., Plaingam, W., Sangsuthum, S., Aizat, W. M., Baharum, S. N., Tencomnao, T., Hu, V. W., et al. (2019). Phenotypic subgrouping and multi-omics analyses reveal reduced diazepam-binding inhibitor (DBI) protein levels in autism spectrum disorder with severe language impairment. PloS one, 14(3).
Pinaya, W. H., Mechelli, A., & Sato, J. R. (2019). Using deep autoencoders to identify abnormal brain structural patterns in neuropsychiatric disorders: A large-scale multi-sample study. Human brain mapping, 40(3), 944–954.
Rapin, I., Dunn, M. A., Allen, D. A., Stevens, M. C., & Fein, D. (2009). Subtypes of language disorders in school-age children with autism. Developmental Neuropsychology, 34(1), 66–84.
Sacco, R., Lenti, C., Saccani, M., Curatolo, P., Manzi, B., Bravaccio, C., & Persico, A. M. (2012). Cluster analysis of autistic patients based on principal pathogenetic components. Autism Research, 5(2), 137–147.
Sevin, J. A., Matson, J. L., Coe, D., Love, S. R., Matese, M. J., & Benavidez, D. A. (1995). Empirically derived subtypes of pervasive developmental disorders: A cluster analytic study. Journal of Autism and Developmental Disorders, 25(6), 561–578.
Shen, J. J., Lee, P. H., Holden, J. J., and Shatkay, H. (2007). Using cluster ensemble and validation to identify subtypes of pervasive devel- opmental disorders. In AMIA Annual Symposium Proceedings, volume 2007, page 666. American Medical Informatics Association.
Silleresi, S., Prevost, P., Zebib, R., Bonnet-Brilhault, F., Conte, D., and Tuller, L. (2020). Identifying language and cognitive profiles in children with ASD via a cluster analysis exploration: Implications for the new ICD-11. Autism Research.
Simpson, K., Adams, D., Alston-Knox, C., Heussler, H. S., & Keen, D. (2019). Exploring the sensory profiles of children on the autism spectrum using the short sensory profile-2 (SSP-2). Journal of autism and developmental disorders, 49(5), 2069–2079.
Smith, A. M., King, J. J., West, P. R., Ludwig, M. A., Donley, E. L., Burrier, R. E., & Amaral, D. G. (2019). Amino acid dysregulation metabotypes: potential biomarkers for diagnosis and individualized treatment for subtypes of autism spectrum disorder. Biological psychiatry, 85(4), 345–354.
Stevens, E., Atchison, A., Stevens, L., Hong, E., Granpeesheh, D., Dixon, D., and Linstead, E. (2017). A cluster analysis of challenging behaviors in autism spectrum disorder. In 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA), pages 661–666. IEEE.
Stevens, E., Dixon, D. R., Novack, M. N., Granpeesheh, D., Smith, T., & Linstead, E. (2019). Identification and analysis of behavioral phenotypes in autism spectrum disorder via unsupervised machine learning. International journal of medical informatics, 129, 29–36.
Stevens, M. C., Fein, D. A., Dunn, M., Allen, D., Wa-terhouse, L. H., Feinstein, C., & Rapin, I. (2000). Subgroups of children with autism by cluster analysis: A longitudinal examination. Journal of the American Academy of Child & Adolescent Psychiatry, 39(3), 346–352.
Tadevosyan-Leyfer, O., Dowd, M., Mankoski, R., Winklosky, B., Putnam, S., McGrath, L., Tager-Flusberg, H., & Fol- stein, S. E. (2003). A principal components analysis of the autism diagnostic interview-revised. Journal of the American Academy of Child & Adolescent Psychiatry, 42(7), 864–872.
Tovar, A. E., Rodriguez-Granados, A., & Arias-Trejo, N. (2020). Atypical shape bias and categorization in autism: Evidence from children and computational simulations. Developmental Science, 23(2), e12885.
Uljarevic, M., Lane, A., Kelly, A., & Leekam, S. (2016). Sensory subtypes and anxiety in older children and adolescents with autism spectrum disorder. Autism Research, 9(10), 1073–1078.
Veatch, O., Veenstra-VanderWeele, J., Potter, M., Pericak- Vance, M. A., & Haines, J. (2014). Genetically meaningful phenotypic sub- groups in autism spectrum disorders. Genes, Brain and Behavior, 13(3), 276–285.
Wiggins, J. L., Peltier, S. J., Ashinoff, S., Weng, S.-J., Carrasco, M., Welsh, R. C., Lord, C., & Monk, C. S. (2011). Using a self- organizing map algorithm to detect age-related changes in functional connec-tivity during rest in autism spectrum disorders. Brain research, 1380, 187–197.
Zheng, L., Grove, R., & Eapen, V. (2019). Spectrum or subtypes? a latent profile analysis of restricted and repetitive behaviours in autism. Research in Autism Spectrum Disorders, 57, 46–54.
Zheng, S., Hume, K. A., Able, H., Bishop, S. L., and Boyd, B. A. (2020). Exploring developmental and behavioral heterogeneity among preschoolers with ASD: A cluster analysis on principal components. Autism Research.
MacQueen, J. (1967, June). Some methods for classification and analysis of multivariate observations. In Proceedings of the fifth Berkeley symposium on mathematical statistics and probability (Vol. 1, No. 14, pp. 281–297).
Lane, Alison E.., Dennis, Simon J.., & Geraghty, Maureen E.. (2011). Brief Report: Further Evidence of Sensory Subtypes in Autism. Journal of Autism and Developmental Disorders, 41(6), 826–831. https://doi.org/10.1007/s10803-010-1103-y.
Zheng, L., Grove, R., & Eapen, V. (2019). Spectrum or subtypes? A latent profile analysis of restricted and repetitive behaviours in autism. Research in Autism Spectrum Disorders, 57, 46–54.
Wiggins, Jillian Lee, Peltier, Scott J.., Ashinoff, Samantha, Weng, Shih-Jen., Carrasco, Melisa, Welsh, Robert C.., et al. (2011). Using a self-organizing map algorithm to detect age-related changes in functional connectivity during rest in autism spectrum disorders. Brain Research, 1380, 187–197. https://doi.org/10.1016/j.brainres.2010.10.102.
Fein, D., Stevens, M., Dunn, M., Waterhouse, L., Allen, D., Rapin, I., & Feinstein, C. (1999). Subtypes of pervasive developmental disorder: Clinical characteristics. Child Neuropsychology, 5(1), 1–23.
Funding
CPP was funded by NSF GRFP, Fellowship ID #1849569.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Parlett-Pelleriti, C.M., Stevens, E., Dixon, D. et al. Applications of Unsupervised Machine Learning in Autism Spectrum Disorder Research: a Review. Rev J Autism Dev Disord 10, 406–421 (2023). https://doi.org/10.1007/s40489-021-00299-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40489-021-00299-y