Abstract
Background
Severe allergic asthma (SAA) is based on type 2 (T2-high) immune responses to allergens promoting type 2 T helper (Th2) cell cytokine responses and production of IgE antibodies. Omalizumab was the first biological drug licensed for clinical use in the management of IgE-mediated SAA. Despite emerging evidence supporting the prominent role of follicular T cells (Tfh), Breg and Treg subsets, in the development and progression of SAA, no data are available on the impact of omalizumab therapy.
Methods
Ten SAA patients monitored at the Respiratory Diseases Unit of Siena University Hospital and ten healthy sex- and age-matched controls were enrolled in the study. Clinical and functional parameters were collected at baseline (T0) and after 6 months of therapy (T6). Cellular population analysis was determined through multicolour flow cytometry.
Results
SAA patients showed higher percentages of Th17.1, Tfh and Tfh2 while CD24hiCD27hi Breg cell, Treg and Tfr percentages were significantly lower than in controls. Higher percentages of Tfh2 in patients with nasal polyps than in those without and in controls were observed. At T6, significant decreases in Tfh and Tfh2 compared with T0 were observed. A slightly significant increase in Teffs was reported at T6 compared to T0. ΔIgE levels in serum were correlated with ΔCD19+CD24+CD27+ Breg cell percentages (r = − 0.86, p = 0.0022).
Conclusions
Our data explored the changes in Tfh cells, Tregs and Bregs in severe asthma. The restoration of immunological imbalance in SAA patients after omalizumab is certainly intriguing and represents a glimpse into the comprehension of immunological effects of treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Our results evidenced a large spectrum of immune change induced by omalizumab, highlighting new perspectives for its therapeutic implications. |
The increase in Tfh2 percentages and the decrease in CD19+CD24hiCD27+ Breg cells in SAA patients were significantly rebalanced after omalizumab therapy. |
CD19+CD24hiCD27+ Breg cells after omalizumab treatment appeared to be strongly correlated with baseline serum concentrations of IgE, suggesting that B-cell imbalance may directly contribute to overproduction of IgE. |
Nasal polyposis with concomitant severe asthma was associated with a significantly higher expression of Tfh2 cells. |
1 Introduction
Severe allergic asthma (SAA) is very difficult to manage in many individuals, and systemic corticosteroids are often used to prevent or manage acute exacerbations
From the immunological point of view, SAA is based on type 2 (T2-high) immune responses to allergens promoting type 2 T-helper (Th2) cell cytokine responses and production of IgE antibodies [1]. There are few data currently available about the expression of specific peripheral B- and T-cell markers in SAA [2,3,4].
Newly discovered CD4+ T-cell subsets, known as follicular helper T (Tfh) cells, in human peripheral blood control IgE production better than do Th2 cells [2]. According to their CXCR3 and CC chemokine receptor type 6 (CCR6) expression profiles, Tfh cells can be clustered as Tfh1 (CXCR3+CCR6−), Tfh2 (CXCR3−CCR6−) and Tfh17 cells (CXCR3−CCR6+) [5]. By balancing inflammatory effects of these cell phenotypes, regulatory cells also play a key role in the development of asthma.
In addition to inflammatory cells, regulatory T cells (Tregs) were observed to play a key role in the development of asthma. Treg cell populations control immune homeostasis, suppress allergic responses, and participate in the resolution of inflammation-associated tissue injuries [6,7,8,9].
A population of regulatory cells, expressing CXCR5+ T, with different functions and activity has also been identified, and is known as T follicular regulatory (Tfr) cells [10].
An increasing number of papers describe a new phenotype of B cells with suppressive function, known as regulatory B cells (Bregs) [11]. Bregs have immunosuppressive activity mediated by IL-10, IL-35 and TGF-beta release, inducing activation of Tregs [12,13,14,15]. Types of human Breg cells have different surface markers characterising different subsets, such as CD27+CD24hi cells, CD24hiCD38hi immature/transitional B cells and CD5+CD1dhi [16].
The absence or loss of Bregs is reported to enhance symptoms and the frequency of exacerbations in patients with allergic diseases [17].
Despite emerging evidence supporting the prominent role of these cell subsets in the development and progression of SAA, no data are available on the impact of anti-IgE biological therapy in these patients.
Omalizumab was the first biological drug licensed for clinical use in the management of IgE-mediated SAA [18]. Available since 2003, it is a recombinant humanized monoclonal antibody that inhibits IgE-dependent cellular and molecular events, involved in the immune/inflammatory cascade, by interaction with high- and low-affinity membrane receptors (FcεRI and FcεRII/CD23) [19]. Although the clinical efficacy of the drug has been widely investigated, cellular immune responses are not fully understood.
The aim of this study was to investigate immunological responses mediated by Tfh, Treg and B regulatory cells in responder SAA patients before and after omalizumab treatment in order to contribute to the definition of the effects of anti-IgE biological therapy on adaptive and innate immunity.
2 Materials and Methods
2.1 Study Population
Fifty-seven SAA patients (median age 53.3 ± 14.7; 38% males), followed at the Respiratory Diseases Unit of Siena University Hospital were included in the study from January 2019 to January 2020. Among these, 46 gave their written informed consent to participate in the study, which was approved by our Local Ethics Committee CEAVSE (code number 180712 and Markerlung 17431). Among these patients, 36 were excluded for different reasons: 28 reported concomitant diseases that can alter immunological data (ten were active smokers, seven had chronic obstructive pulmonary disease, four had autoimmune disorders, one had cancer, and six were diabetics), while eight patients were discontinued on omalizumab therapy (three for adverse reactions and one for a hypersensitivity reaction to omalizumab, three for non-adherence to the treatment, and one due to death in a car accident).
Ten SAA patients (median age 54.6 ± 7.5; 30% males) and ten healthy sex- and age-matched controls were finally included in the study.
SAA was diagnosed according to the Global Strategy for Asthma Management and Prevention, Global Initiative for Asthma (GINA) guidelines [20]. The results of the asthma control questionnaire (ACQ) and the asthma control test (ACT), physical examination and lung function tests at baseline and during follow-up were collected in a database. All SAA patients included in the study were treated with subcutaneous injections of omalizumab: dose and frequency of injections were calculated according to body weight and total serum IgE. Serum levels of IgE-specific allergens and nonspecific (total) IgE were measured by Immuno-CAP 250 (Thermo Fisher Scientific, Waltham, MA, USA), a highly automated system, requiring minimal hands-on time. The results were expressed as U/mL. Serum and blood sampling were obtained at baseline and after 6 months (T6) of therapy with omalizumab. The ten healthy controls (four male, age 48 ± 12 years) were normal on physical examination, and were all negative to the Immuno-CAP test. They had no history of asthma or allergy and were not on any medication. They were monitored for at least 12 months and did not develop any disease.
2.2 Lung Function Tests
The following lung function parameters were recorded according to ATS/ERS standards using a Jaeger body plethysmograph with corrections for temperature and barometric pressure: forced vital capacity (FVC), forced expiratory volume in the first second (FEV1), and FEV1/FVC ratio. All were expressed as percentages of predicted values. Lung function tests were performed at baseline and at follow-up.
2.3 Antibodies
Multicolour flow cytometric analysis was performed using the following fluorochrome-labelled anti-human monoclonal antibodies for measuring lymphocyte Th and Tfh subsets (CD4-APC-Vio770, CD45RA-PE-Vio770, CCR6-APC, CXCR3-VioBright FITC CXCR5 PerCP-Cy5.5, CCR4-PE) all from Miltenyi biotech. Tregs and Tfr were detected using BD Human Regulatory T Cell Cocktail (BD, San Jose, CA, USA), including CD4-FITC (clone SK3), CD25-PECy7 (clone 2A3), CD127 Alexa Fluor® 647 and adding CXCR5 PerCP-Cy5 and CD8 horizon450 (the last two from Biolegend). Bregs were detected using the following fluorochrome-labelled anti-human monoclonal antibodies: CD38-FITC CD1d-PE, CD19-PE-Cy7, CD5-PerCP-CY5.5, CD24-APC-H7 and CD27- BV510, all from BD Biosciences (San Jose, CA, USA).
2.4 Preparation and Storage of Peripheral Blood Mononuclear Cells
Preparation and storage of peripheral blood mononuclear cells (PBMCs) was performed in the laboratory of the Respiratory Diseases Unit, Siena University Hospital (Italy) from January 2018 to June 2020. Peripheral blood samples after 8 h of fasting were drawn into a tube containing EDTA anticoagulant (BD Vacutainer® EDTA tubes, BD Biosciences) and processed within 8 h.
Briefly, a layer of blood was added to the same volume of Ficoll Histopaque®-1077 (Sigma-Aldrich, Burlington, MA, USA) in a 50 mL tube and centrifuged for 30 min at 1308g in a swinging-bucket rotor without brake. The mononuclear cell layer was transferred to a new conical tube (Corning® 50 mL centrifuge tubes, Sigma-Aldrich), adding 15 mL RPMI 1640 medium (Gibco®, Thermo Fisher Scientific), and centrifuged at 327g for 10 min. Supernatant was carefully removed and after being kept for 1 h at − 20 °C and overnight at − 80 °C, the cells were stored in liquid nitrogen until analysis.
The vitality and the number were defined through trypan blue chamber and the correct amount of cells were used for cell staining.
Cell staining and flow cytometric analysis were performed using FACS CantoII cell analyser (BD Biosciences). Briefly, the cells were thawed and centrifuged at 327g for 10 min, discarding the supernatant. A total of 106 live cells were added to each tube (based on trypan-blue stain detection in a Burker chamber). Cells were marked with an appropriate amount of mABs and incubated for 30 min, according to the manufacturer's instructions. After incubation, the cells were resuspended in medium and read by flow cytometer. Kaluza Analysis 2.1 software was used for data analysis.
2.5 Flow Cytometric Analysis
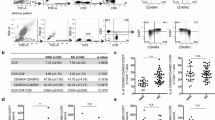
We used a FACS CantoII (BD Biosciences) cell analyser and Kaluza Analysis 2.1 software (Beckman and Coulter). The first tube contained the antibodies for analysis of Th and Tfh subsets. T cells were selected in the lymphocyte scatter region (gating out dead cells that show low forward and relatively high side scatter) and then gated for the CD4 phenotype. CD4+ cells were clustered based on CD45RA and CCR6 expression. In particular, total CD4+ cells were divided into CD4+CD45RA+ cells (Teffs) and CD4+CD45RA− T central memory cells (Tcms). Tcms were subsequently divided according to CCR6 expression. CD4+CD45RA−CCR6+ cells were classified as CXCR3+CCR4+ (Th17 double-positive), CXCR3−CCR4+ (Th17) and CXCR3+CCR4− (Th1-like Th17), whereas CCR6−cells were classified as CXCR3−CCR4+ (Th2) and CXCR3+CCR4− (Th1). CD4+CD45RA− cells were also used for analysis of Tfh. In particular, CD4+CD45RA−CXCR5+ were considered to be Tfh and their subsets were identified: CXCR3−CCR6+ Tfh17, CXCR3+CCR6− (Th1) and CXCR3−CCR6− (Tfh2). Gate strategy was performed according to Miyajima et al. [21], and is reported in Fig. 1a. The second tube contained the antibodies for analysis of Tregs and Tfr. T cells were selected in the lymphocyte scatter region (gating out dead cells that show low forward and relatively high side scatter), then gating for CD4 cells. We then made a plot for the identification of CD25highCD127low Tregs. An additional histogram was plotted for CXCR5+ CD25highCD127low Tfr. Gate strategy is reported in Fig. 2a [22]. The third tube contained the antibodies for analysis of Bregs. B cells were gated on the basis of CD19 phenotype. Three additional dot plots were added to recognize the following B-cell subsets: CD24hiCD38hi Breg, CD24hiCD27hi Breg and CD5+CD1d+ Breg. Gate strategy is reported in Fig. 3a [23].
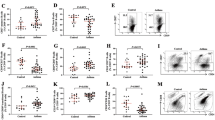
a Gating strategy used to analyse markers of differentiation of T-cell subsets. They are identified as CD4+CD45RA+ Cells (Teff) and CD4+CD45RA- T central memory cells (Tcms). Tcms were subsequently divided according to expression of CCR6. CD4+CD45RA-CCR6+ cells were classified as CXCR3+CCR4+ (Th17 double-positive), CXCR3-CCR4+ (Th17) and CXCR3+CCR4- (Th1-like Th17), whereas CCR6- cells were classified as CXCR3-CCR4+ (Th2) and CXCR3+CCR4- (Th1). CD4+CD45- cells were also used for the analysis of Tfh. In particular CD4+CD45RACXCR5+ were considered as Tfh and CXCR3-CCR6+ Tfh17 and CXCR3+CCR6- Tfh1, CXCR3-CCR6- Tfh2. b, c Percentages of different Tcm, Teff Tfh, Tfh2 and Th17.1 cell subpopulations in controls (n = 10), patients at baseline (n = 10) (b), and T6 (n = 10) (c) obtained by manual gating. Data represent individual values, mean (centre bar) ± SEM (upper and lower bars). CI confidence interval. Mann-Whitney U test and Wilcoxon ranked test were used. p < 0.05 was used for statistical significance
a Gating strategy used to analyse markers of differentiation of regulatory T-cell subsets. They are identified as CD4+ CD25highCD127 low Treg and CD25 high CD127 low CXCR5+ Tfr. b, c Percentages of different Tfr and Treg cell subpopulations in controls (n = 10), patients at baseline (n = 10), and T6 (n = 10) obtained by manual gating. Data represent individual values, mean (centre bar) ± SEM (upper and lower bars). CI confidence interval. Mann-Whitney U test and Wilcoxon ranked test were used. p < 0.05 was used for statistical significance
a Gating strategy used to analyse markers of differentiation of B regulatory cells. They are identified as CD19+CD24+CD27+ Breg, CD19+CD24+CD38+Breg and CD19+CD1d+CD5+ Breg. b, c Percentages of different CD19+CD24+CD27+ Breg cell subpopulations in controls (n = 10) and patients at baseline (n = 10), obtained by manual gating. Data represent individual values, mean (centre bar) ± SEM (upper and lower bars). CI confidence interval. Mann-Whitney U test and Wilcoxon ranked test were used. p < 0.05 was used for statistical significance
2.6 Statistical Analysis
All data are reported as median and interquartile range (IQR) or mean and standard error of the mean (M ± SEM). Significant differences between controls and patients at baseline were determined by Student’s t-test or the Mann-Whitney U test. Wilcoxon’s rank sum test was used to compare data before and after treatment (T0–T6). Correlations were determined by Spearman correlation coefficient. Probability values less than 0.05 were considered significant. Statistical analysis was performed by GraphPad Prism 8.0 software. Unsupervised principal component analysis (PCA) was employed to reduce the dimensionality of data hyperspace. The data matrix was constructed with Microsoft Excel and PCA was performed using BioVinci software (BioTuring, San Diego, CA, USA). The cell subsets of patients and controls were also used to build a decision-tree model to determine the best clustering variables. The cellular subsets of patients were also employed to create a decision tree model for the detection of best clustering variables through the Gini criterion.
3 Results
3.1 Study Population and Clinical Improvements After 6 Months of Therapy
The main characteristics of our study population are reported in Table 1 and Supplementary Table 2 (Online Supplementary Material, OSM), including IgE assessment, clinical data and therapeutic status.
In terms of clinical asthma control, a mean of 3.6 ± 2 exacerbations occurred in the year prior to administration of omalizumab.
After 6 months of omalizumab therapy, we observed a significant increase in FEV1% (75 ± 7 vs. 87.8 ± 5.3; p = 0.002), FVC/FEV1 (68 ± 4 vs. 73 ± 4.6; p = 0.002), and an improvement in clinical asthma control, expressed by the ACT score (13.5 ± 1.6 vs. 21.7 ± 1; p = 0.002). On the contrary, a significant decrease in total IgE concentrations in serum was reported (Fig. 4) (425 ± 116 vs. 116 ± 25 U/mL; p = 0.004).
Therapy with omalizumab was not associated with any moderate-to-severe adverse events and there were no interruptions of treatment.
3.2 T-Cell Subsets in Severe Allergic Asthma (SAA) Patients Before Treatment and Controls
Immunological cell percentages are reported in Supplementary Table 1 (OSM). Th and Tfh cells, analysed in SAA patients at baseline and in controls (Fig. 1b), showed higher percentages of Tcms (70.2 ± 2.8% vs. 44.13 ± 4%, p < 0.0001), Th17.1 (29.6 ± 4.2% vs. 17.6 ± 2.7%; p = 0.04), Tfh (14.2 ± 2.5% vs. 5.5 ± 1.2%; p = 0.0037) and Tfh2 (43.5 ± 8.9% vs. 34.3 ± 1.6%; p = 0.02) in patients than in controls. Teffs were lower in patients than in controls (28.9 ± 2.6% vs. 53.1 ± 1.9%; p = 0.001).
Stratifying our cohort for patients with and without nasal polyps, we recorded higher percentages of Tfh2 at baseline in patients with nasal polyps than in those without and in controls (65.9 ± 9.5 vs. 28.8 ± 98 and 34.3 ± 1.6%; p = 0.03).
Interestingly, Treg and Tfr percentages (Fig. 2b) were significantly lower in patients than in controls (3.2 ± 0.6% vs. 7.2 ± 1.4%; p = 0.019 and 0.5 ± 0.2% vs. 3.1 ± 0.4; p = 0.0001).
Regarding Breg cell analysis (Fig. 3b), CD24hiCD27hi Breg cell percentages were significantly lower in patients than in controls (7 ± 0.9% vs. 14.7 ± 4%; p = 0.01).
3.3 T-Cell Subsets in SAA Patients Before and After Therapy
At T6, we observed a significant decrease in Tcms (56.6 ± 6.6% vs. 70.2 ± 2.8; p = 0.048), Tfh (7.4 ± 1.6% vs. 14.2 ± 2.5%; p = 0.003) and Tfh2 (35.5 ± 8.6% vs. 43.5 ± 8.9%; p = 0.03). A slightly significant increase in Teffs (43.2 ± 6.6% vs. 28.9 ± 2.6; p = 0.04) was reported at T6 with respect to T0 (Fig. 1c). A decrease in Th17.1 percentage was recorded but was not statistically significant (24 ± 3.5% vs. 29.6 ± 4.2%; p = 0.08).
Stratifying our cohort for nasal polyposis, a significant decrease in Tfh2 percentage was only found in patients with nasal polyposis at T6 (65.9 ± 9.5 vs. 48.8 ± 33%; p = 0.007).
Treg and Tfr percentages showed a significant increase at T6 (3.2 ± 0.6% vs. 7 ± 1.8%; p = 0.02, and 0.5 ± 0.2% vs. 1.8 ± 0.3; p = 0.002, respectively (Fig. 2c), as CD24hiCD27hi Breg cells (7 ± 0.9% vs. 12.6 ± 2.5%; p = 0.01; Fig. 3c).
3.4 Correlation Analysis
At baseline, total IgE levels in serum were correlated with ACT score (r = − 0.67, p = 0.036), body mass index (BMI) (r = 0.63, p = 0.04) and Tfh1 (r = 0.72, p = 0.02). BMI showed an indirect correlation with ACT score (r = − 0.69, p = 0.030) and Tfh17 (r = − 0.78, p = 0.01) and a direct correlation with Tfh1 (r = 0.81, p = 0.005).
ACT score was correlated with Treg (r = 0.73, p = 0.01), Tfh2 (r = − 0.81, p = 0.0064), Tfh1 (r = − 0.77, p = 0.01) and Tfh17 (r = 0.87, p = 0.0015) percentages.
Among cell phenotypes, Th2 was correlated with Th17 (r = 0.68, p = 0.03), Th1(r = − 0.84, p = 0.004) and CD38+CD24+ Breg cell percentages (r = − 0.77, p = 0.013). CD38+CD24+ Breg cell percentages were indirectly correlated with Tfh cell percentages (r = − 0.58, p = 0.04). Th17 cell percentages were correlated with those of CD19+ B cells (r = 0.7, p = 0.007). CD8+ cell percentages were correlated with Tfh cell subsets (Tfh1 r = 0.74, p = 0.017; Tfh2 r = 0.75, p = 0.015; and Tfh17 r = − 0.75, p = 0.015).
The number of peripheral eosinophils (cells/mL) at baseline was correlated with Th17.1 cell percentages (r = − 0.90, p = 0.0046) and number of exacerbations in the year prior to treatment (r = 0.90, p = 0.004).
Regarding the variation in variables between T6 and T0, Δ(T6–T0) was calculated, following by correlation analysis.
Interestingly, ΔIgE levels in serum were correlated with ΔCD19+CD24+CD27+ Breg cell percentages (r = − 0.86, p = 0.0022; Fig. 5a), and ΔFEV1% (r = − 0.91, p = 0.0005). Moreover, ΔCD19+CD24+CD27+ Breg cell percentages directly correlated with ΔFEV1% (r = 0.74, p = 0.01, Fig. 5b).
ΔFEV1 (mL) inversely correlated with ΔTfh2 (r = − 0.78, p = 0.0097).
3.5 Multivariate Analysis
Analysis of variance of cell populations in controls showed a homogeneous distribution with low interindividual variance (red boxes) (Fig. 6a) for many of the 22 cell populations analysed. When SAA patient cell populations were compared at baseline and at T6, significant differences expressed as high interindividual variance (green boxes) were found for many of the 22 populations analysed.
a Interindividual variance (CV) for 22 canonical cell populations was calculated for each three conditions for T0, T6 (6 months) and HC samples. b Principal component analysis (PCA) of T cell. c The 22 cellular subsets of patients and controls were employed to create a decision tree model for the detection of best clusterising variables
PCA plots were performed in order to distinguish the three groups: HC, T0 and T6. The analysis shows how the three groups (controls, T0 and T6) were separate on the basis of cell phenotype (Fig. 6b). The first and second components explained 35.34% and 27.70% of the total variance.
The cell subsets of patients and controls were employed to build a decision-tree model to determine the best clustering variables (Fig. 6c). The best cell populations were Teff followed by Tregs, Th17.1, CD19+CD24+CD27+ Breg and CD5+CD1d+ Breg cells.
4 Discussion
We studied the expression of Th, Tfh and Breg cells and their subsets for the first time in SAA patients before and during treatment with omalizumab. Compared with the group of healthy controls, we observed a significant imbalance of these immunological cell subtypes in SAA patients: a reduction in Tregs and CD19+CD24+CD27+ Breg cells and overexpression of Tfh, Tfh2 and Tcm cells, as previously reported in the literature [24,25,26]. After 6 months of omalizumab treatment, a normalising, even if partial, trend of cell profiles was observed, again for the first time, in SAA patients, associated with clinical improvement and increases in FEV1% and FVC/FEV1 ratio.
Despite the large amount of work assessing the clinical effectiveness of omalizumab in SAA patients, the impact of this biological treatment on the different subtypes immunological cells remains largely unexplored. Gruchalla et al. interestingly investigated the effects of omalizumab on T-lymphocyte function in inner-city children with asthma, reporting no significant alterations on blood T-cell percentages and activation [28]. On the other hand, Rauber et al. showed that omalizumab has an impact on inflammatory T-cell activity, observing a significant correlation between clinical treatment response and reduced numbers of IL-10-, IL-31- and IFN-γ-secreting cells [29].
Moreover, many new studies have emphasised the potentially crucial role of Tfh cells and their subsets in the progression and pathogenesis of allergic asthma. In a mouse model of airway allergen exposure, it was demonstrated that Tfh cells supported sustained production of IgE in vivo [30, 31]. Moreover, Kamekura et al. demonstrated increased percentages of Tfh2 cells in patients with allergic rhinitis with and without asthma and a simultaneous decrease in CD19+CD24+CD27+ Breg cells [32]. Miyajima et al. recently demonstrated that Tfh2 [32] cells were more abundant and that Tfh1 cells and CD19+CD24+CD27+ Breg cells were much sparser in allergic patients than in healthy controls. After inhaled corticosteroid treatment, Tfh2 percentages declined, while symptoms and clinical biomarkers improved, suggesting that the expression of these cell subsets was strictly related to clinical assessment and response to therapy [21]. Our results are in line with these findings, as our patients showed a comprehensive rebalancing of Th and Tfh cell subtypes after a relatively short time of omalizumab treatment, leading to significant clinical improvement.
In the same way, the increase in Tfh2 percentages and the decrease in CD19+CD24hiCD27+ Breg cells in SAA patients were significantly rebalanced after omalizumab therapy, supporting the effectiveness of this drug as an ‘immunological restorer’ in these patients. The immunological mechanisms through which omalizumab was able to induce these modifications need to be clarified. Tfh2 cells and CD19+CD24hiCD27+ Breg cells are not reported to express specific surface receptors for IgE (FcεRI and CD23), suggesting that their restoration after omalizumab treatment is not directly related to IgE blockage, but may be determined by the drug interaction with dendritic cells, which are crucial in the development of Tfh [33, 34]
Gong et al. demonstrated that circulating Tfh cells induce naive and memory B cells to become Ig-producing cells. A high frequency of these cells was reported in patients with asthma [25]. The authors also reported an increase in Tfh cells and accelerated IgE production when co-culture with CD19+ B cells was performed [36]. Abdel-Gadir et al. demonstrated that omalizumab promotes allergen desensitization through initial depletion of allergen-reactive T cells, followed by an increase in allergen-specific Treg cell activity due to reversal of their Th2 cell-like programme [37]. In our cohort, these baseline immunological characteristics were confirmed, showing a significant increase of Breg and Treg cells after omalizumab treatment. Interestingly, the degree of the increase in CD19+CD24+CD27+ Bregs after omalizumab treatment appeared to be strongly correlated with baseline serum concentrations of IgE, suggesting that B-cell imbalance may directly contribute to overproduction of IgE. However, all these changes can be considered as an ‘indirect effect’ of omaliozumab, able to block IgE.
Concerning different clinical phenotypes of SAA, including comorbidities, we observed that nasal polyposis was associated with a significantly higher expression of Tfh2 cells. These findings are in line with previous studies, in which an increase in Tfh and especially Tfh2 cells in nasal polyp tissue was demonstrated after induction of local IgE production [35].
With regard to body mass index (BMI), new evidence has demonstrated the relationship with omalizumab therapy. In particular, Gu et al. found an inverse correlation between BMI and ACT score. On the other hand, IgE levels directly correlate with BMI [27]. In our cohort the same findings with these parameters were reported, suggesting an intriguing role of BMI in omalizumab efficacy.
With regard to Tfr cells, no study has yet addressed the question of involvement of Tfr cells in asthma patients [36]. Our study is the first to show that Tfr concentrations are lower in SAA patients than in controls and are enhanced by anti-IgE treatment, supporting the hypothesis that these cell subsets are involved in the effects of omalizumab therapy. Since Tfr cells are crucial in the downregulation of Tfh and plasma cells activity, our findings are certainly interesting and provide intriguing insights in terms of immunological dysregulation of SAA patients.
Concerning Th17 cells, few and controversial data are available with regard to their role in the pathogenesis of allergic asthma [38]. It has been reported that Th17 cells have phenotypic instability that predisposes them to a pro‐inflammatory phenotype. This plasticity affects Th effector/memory cells [38, 39]. Our results demonstrated that in a population of SAA patients treated with omalizumab, Th17 cells showed inflammatory properties that may coexist with Th2. At baseline, Th2 and Th17 inflammation matched an increase in Tfh and B cells and a depletion of Th1 and CD38+CD24+ Breg cells, while an opposite trend was shown with Tfh17 cells, which decreased when Tfh2, Tfh1 and CD8 cell numbers were enhanced. The number of eosinophils was directly correlated with Th17.1 cells and decreased after omalizumab therapy. The interaction between cell subsets and clinical parameters remained stable after treatment, the proinflammatory effect of Tfh1, Tfh2 and B cells showing an inverse correlation with Tfh17 cells. These results are certainly intriguing and further underline the high complexity of the immunoinflammatory milieu underlying the multiple endotypes of severe asthma.
4.1 Limitations
Our study has several limitations, including the monocentric retrospective design, the relatively short follow-up time for measurement of immunological and clinical data after omalizumab treatment, the small sample size, and the absence of intracellular staining. However, our results provide a comprehensive glance on the immunological impact of omalizumab in SAA patients. A large number of cell subsets were evaluated simultaneously to obtain insights into their complex interactions: our data explored the changes in Tfh cells, Tregs and Bregs in severe asthma during omalizumab treatment. Our results showed a large spectrum of immune change induced by omalizumab, suggesting new perspectives for their therapeutic implications. The overall restoration of immunological imbalance in SAA patients with omalizumab, not only limited to allergic mechanisms, is certainly intriguing and may represent a glimpse of light in the comprehension of the immunological effects of biological treatment in SAA.
Change history
21 December 2021
The original online version of this article was revised in order to add the funding note information
References
Caminati M, Pham DL, Bagnasco D, Canonica GW. Type 2 immunity in asthma. World Allergy Organ J. 2018;11:13. https://doi.org/10.1186/s40413-018-0192-5.
Yao Y, Chen C-L, Yu D, Liu Z. Roles of follicular helper and regulatory T cells in allergic diseases and allergen immunotherapy. Allergy. 2020. https://doi.org/10.1111/all.14639.
Yao Y, Wang N, Chen C-L, Pan L, Wang Z-C, Yunis J, Chen Z-A, Zhang Y, Hu S-T, Xu X-Y, Zhu R-F, Yu D, Liu Z. CD23 expression on switched memory B cells bridges T-B cell interaction in allergic rhinitis. Allergy. 2020;75:2599–612. https://doi.org/10.1111/all.14288.
Cameli P, Bergantini L, d’Alessandro M, Perruzza M, Cekorja B, Perillo F, Massa E, Ruzza A, Fossi A, Beltrami V, Sestini P, Bargagli F (2020) A Comprehensive Evaluation of Mepolizumab Effectiveness in a Real-Life Setting. International Archives of Allergy and Immunology 181(8):606-612. https://doi.org/10.1159/000507996
Berghi NO, Dumitru M, Vrinceanu D, Ciuluvica RC, Simioniuc-Petrescu A, Caragheorgheopol R, Tucureanu C, Cornateanu RS, Giurcaneanu C. Relationship between chemokines and T lymphocytes in the context of respiratory allergies (review). Exp Ther Med. 2020;20:2352–60. https://doi.org/10.3892/etm.2020.8961.
Khan MA. Regulatory T cells mediated immunomodulation during asthma: a therapeutic standpoint. J Transl Med. 2020;18:456. https://doi.org/10.1186/s12967-020-02632-1.
Bergantini L, Cameli P, d’Alessandro M, Vietri L, Perruzza M, Pieroni M, Lanzarone N, Refini R, Fossi A, Bargagli E. Regulatory T cells in severe persistent asthma in the era of monoclonal antibodies target therapies. Inflammation. 2020;43:393–400. https://doi.org/10.1007/s10753-019-01157-0.
Vantaggiato L, Perruzza Rosa M, Refini M, Bergantini L, d’Alessandro M, Cameli P, Perruzza D, Bini L, Bargagli E, Landi C (2020) Mepolizumab and Benralizumab in Severe Eosinophilic Asthma: Preliminary Results of a Proteomic Study. Lung 198(5):761-765. https://doi.org/10.1007/s00408-020-00379-6
Landi C, Cameli P, Vantaggiato L, Bergantini L, d'Alessandro M, Perruzza M, Carleo A, Shaba E, Di Giuseppe F, Angelucci S, Bargagli E, Bini L (2021) Ceruloplasmin and oxidative stress in severe eosinophilic asthma patients treated with Mepolizumab and Benralizumab. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1869(2):140563. https://doi.org/10.1016/j.bbapap.2020.140563
Xie MM, Dent AL. Unexpected help: follicular regulatory T cells in the germinal center. Front Immunol. 2018. https://doi.org/10.3389/fimmu.2018.01536.
Gao X, Ren X, Wang Q, Yang Z, Li Y, Su Z, Li J. Critical roles of regulatory B and T cells in helminth parasite-induced protection against allergic airway inflammation. Clin Exp Immunol. 2019;198:390–402. https://doi.org/10.1111/cei.13362.
Kessel A, Haj T, Peri R, Snir A, Melamed D, Sabo E, Toubi E. Human CD19(+)CD25(high) B regulatory cells suppress proliferation of CD4(+) T cells and enhance Foxp3 and CTLA-4 expression in T-regulatory cells. Autoimmun Rev. 2012;11:670–7. https://doi.org/10.1016/j.autrev.2011.11.018.
Ma S, Satitsuksanoa P, Jansen K, Cevhertas L, van de Veen W, Akdis M. B regulatory cells in allergy. Immunol Rev. 2020. https://doi.org/10.1111/imr.12937.
Bergantini L, d’Alessandro M, Cameli P, Bianchi F, Sestini P, Bargagli Rosa E, Refini M (2020) Personalized Approach of Severe Eosinophilic Asthma Patients Treated with Mepolizumab and Benralizumab. International Archives of Allergy and Immunology 181(10) 746-753. https://doi.org/10.1159/000508936
Bergantini L, d’Alessandro M, Cameli P, Bono C, Perruzza M, Biagini M, Pini L, Bigliazzi C, Sestini P, Dotta F, Bargagli E (2021) Regulatory T cell monitoring in severe eosinophilic asthma patients treated with mepolizumab. Scandinavian Journal of Immunology 94(1). https://doi.org/10.1111/sji.13031
van de Veen W, Stanic B, Wirz OF, Jansen K, Globinska A, Akdis M. Role of regulatory B cells in immune tolerance to allergens and beyond. J Allergy Clin Immunol. 2016;138:654–65. https://doi.org/10.1016/j.jaci.2016.07.006.
Satitsuksanoa P, van de Veen W, Akdis M. B-cell responses in allergen immunotherapy. Curr Opin Allergy Clin Immunol. 2019;19:632–9. https://doi.org/10.1097/ACI.0000000000000580.
Cameli P, D’Alessandro M, Bergantini L, Silvestri E, Romaldi A, Emmi G, Parronchi P, Bargagli E. A multicenter approach to evaluate omalizumab effectiveness in Samter’s triad. Monaldi Arch Chest Dis. 2020. https://doi.org/10.4081/monaldi.2020.1397.
Pelaia C, Calabrese C, Terracciano R, de Blasio F, Vatrella A, Pelaia G. Omalizumab, the first available antibody for biological treatment of severe asthma: more than a decade of real-life effectiveness. Ther Adv Respir Dis. 2018;12:175346661881019. https://doi.org/10.1177/1753466618810192.
GINA-2019-main-report-June-2019-wms.pdf. https://ginasthma.org/wp-content/uploads/2019/06/GINA-2019-main-report-June-2019-wms.pdf. Accessed 22 Jan 2021
Miyajima S, Shigehara K, Kamekura R, Takaki H, Yabe H, Ikegami I, Asai Y, Nishikiori H, Chiba H, Uno E, Takahashi H, Ichimiya S. Activated circulating T follicular helper cells and skewing of T follicular helper 2 cells are down-regulated by treatment including an inhaled corticosteroid in patients with allergic asthma. Allergol Int. 2020;69:66–77. https://doi.org/10.1016/j.alit.2019.08.008.
Mahnke YD, Beddall MH, Roederer M. OMIP-017: Human CD4+Helper T-cell subsets including follicular helper cells. Cytometry A. 2013;83:439–40. https://doi.org/10.1002/cyto.a.22269.
Liechti T, Günthard HF, Trkola A. OMIP-047: High-dimensional phenotypic characterization of B cells. Cytometry A. 2018;93:592–6. https://doi.org/10.1002/cyto.a.23488.
Habener A, Happle C, Skuljec J, Busse M, Grychtol R, Obernolte H, Braun A, Sewald K, Meyer-Bahlburg A, Hansen G. B cells are crucial in the regulation of airway hyperreactivity in an experimental model of asthma. Eur Respir J. 2019. https://doi.org/10.1183/13993003.congress-2019.PA4362.
Gong F, Su Q, Jiang D, Chen J, Pan Y, Huang X. High frequency of circulating follicular helper T cells in patients with bronchial asthma. Clin Lab. 2014;60:963–8. https://doi.org/10.7754/clin.lab.2013.130427.
Yao Y, Wang Z-C, Yu D, Liu Z. Role of allergen-specific T-follicular helper cells in immunotherapy. Curr Opin Allergy Clin Immunol. 2018;18:495–501. https://doi.org/10.1097/ACI.0000000000000480.
Gu C, Upchurch K, Mamaril-Davis J, Wiest M, Lanier B, Millard M, Turner J, Joo H, Oh S. Obesity influences the outcomes of anti-IgE (omalizumab) therapy of asthma. Clin Exp Allergy. 2020;50:1196–9. https://doi.org/10.1111/cea.13696.
Gruchalla RS, Sampson HA, Liu AH, Shreffler W, Wallace PK, Togias A, David G, Calatroni A, LeBeau P. Effects of omalizumab on T lymphocyte function in inner-city children with asthma. Pediatr Allergy Immunol. 2016;27:328–31. https://doi.org/10.1111/pai.12508.
Rauber MM, Pickert J, Holiangu L, Möbs C, Pfützner W. Omalizumab response correlates with reduced IFN-γ-, IL-10- and IL-31-secreting cells in chronic spontaneous urticaria. J Eur Acad Dermatol Venereol. 2020;34:2078–85. https://doi.org/10.1111/jdv.16216.
Noble A, Zhao J. Follicular helper T cells are responsible for IgE responses to Der p 1 following house dust mite sensitization in mice. Clin Exp Allergy. 2016;46:1075–82. https://doi.org/10.1111/cea.12750.
Kobayashi T, Iijima K, Dent AL, Kita H. Follicular helper T cells mediate IgE antibody response to airborne allergens. J Allergy Clin Immunol. 2017;139:300-313.e7. https://doi.org/10.1016/j.jaci.2016.04.021.
Kamekura R, Shigehara K, Miyajima S, Jitsukawa S, Kawata K, Yamashita K, Nagaya T, Kumagai A, Sato A, Matsumiya H, Ogasawara N, Seki N, Takano K, Kokai Y, Takahashi H, Himi T, Ichimiya S. Alteration of circulating type 2 follicular helper T cells and regulatory B cells underlies the comorbid association of allergic rhinitis with bronchial asthma. Clin Immunol. 2015;158:204–11. https://doi.org/10.1016/j.clim.2015.02.016.
Gao N, Dresel J, Eckstein V, Gellert R, Störch H, Venigalla RKC, Schwenger V, Max R, Blank N, Lorenz H-M, Tretter T. Impaired suppressive capacity of activation-induced regulatory B cells in systemic lupus erythematosus. Arthritis Rheumatol. 2014;66:2849–61. https://doi.org/10.1002/art.38742.
Flores-Borja F, Bosma A, Ng D, Reddy V, Ehrenstein MR, Isenberg DA, Mauri C. CD19+CD24hiCD38hi B cells maintain regulatory T cells while limiting TH1 and TH17 differentiation. Sci Transl Med. 2013;5:173ra23. https://doi.org/10.1126/scitranslmed.3005407.
Zhang Y-N, Song J, Wang H, Wang H, Zeng M, Zhai G-T, Ma J, Li Z-Y, Liao B, Wang B-F, Zhen Z, Wang N, Cao P-P, Lin P, Ning Q, Liu Z. Nasal IL-4(+)CXCR5(+)CD4(+) T follicular helper cell counts correlate with local IgE production in eosinophilic nasal polyps. J Allergy Clin Immunol. 2016;137:462–73. https://doi.org/10.1016/j.jaci.2015.07.025.
Gong F, Zhu H-Y, Zhu J, Dong Q-J, Huang X, Jiang D-J. Circulating CXCR5+CD4+ T cells participate in the IgE accumulation in allergic asthma. Immunol Lett. 2018;197:9–14. https://doi.org/10.1016/j.imlet.2018.03.001.
Abdel-Gadir A, Schneider L, Casini A, Charbonnier L-M, Little SV, Harrington T, Umetsu DT, Rachid R, Chatila TA. Oral immunotherapy with omalizumab reverses the Th2 cell-like programme of regulatory T cells and restores their function. Clin Exp Allergy. 2018;48:825–36. https://doi.org/10.1111/cea.13161.
Hynes GM, Hinks TSC. The role of interleukin-17 in asthma: a protective response? ERJ Open Res. 2020. https://doi.org/10.1183/23120541.00364-2019.
Bhaumik S, Basu R. Cellular and molecular dynamics of Th17 differentiation and its developmental plasticity in the intestinal immune response. Front Immunol. 2017;8:254. https://doi.org/10.3389/fimmu.2017.00254.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Acknowledgements
None.
Funding and conflict of interest
This research did not receive any specific funding from agencies in the public, commercial or non-profit sectors.
Funding
Open access funding provided by Università degli Studi di Sienawithin the CRUI-CARE Agreement.
Statement of Ethics
All subjects gave their written informed consent to participate in the study.
Ethical approval
The study was approved by Local Ethics Committee CEAVSE (code number 180712 and Markerlung 17431).
Data availability statement
All data generated or analysed during this study are included in this published article (and its Online Supplementary files).
Disclosure Statement
The authors have no conflicts of interest to declare.
CRediT taxonomy
Conceptualization: LB, MD; methodology: PC, TP; formal analysis and investigation: PC, MF; writing – original draft preparation: LB, Md; writing – review and editing: EB, PS; supervision: EB.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by PC, TP and MF. The first draft of the manuscript was written by LB and MD; all authors commented on previous versions of the manuscript. EB and PS supervised the manuscript. All authors read and approved the final manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Bergantini, ., d’Alessandro, ., Cameli, . et al. Follicular T Helper and Breg Cell Balance in Severe Allergic Asthma Before and After Omalizumab Therapy. Mol Diagn Ther 25, 593–605 (2021). https://doi.org/10.1007/s40291-021-00545-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-021-00545-3