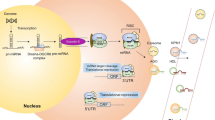
Abstract
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression, involved in the silencing of messenger RNA (mRNA) translation. The importance of miRNA signatures in disease screening, prognosis, and progression of different tumor types and subtypes is increasing. miRNA expression levels change depending on numerous factors. In this review, we are describing the circumstances under which miRNA levels can change, these are named ‘levels’ of heterogeneity of miRNAs. miRNAs can have oncogenic, tumor suppressive, or both roles depending on tumor type and target mRNA whose translation they silence. The expression levels of a single miRNA may vary across different cancer types and subtypes, indicating that a miRNA signature may be tissue specific. miRNA levels of expression also vary during disease formation and propagation, indicating the presence of a time profile for their expression. The complexity of the miRNA-mRNA interference network mirrors different genetic and epigenetic changes that influence miRNA and mRNA availability to each other, and hence, their binding ability. The potential role of miRNAs as biomarkers is two-fold; first, for monitoring of the phases of cancer pathogenesis, and second, to characterize the particular type/subtype of cancer. It is important that a particular miRNA should be characterized by examining as many types and subtypes of cancers as are available, as well as being extracted from different types of samples, in order to obtain a complete picture of its behavior and importance in the disease pathology.


Similar content being viewed by others
References
Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–4.
Galasso M, Sandhu SK, Volinia S. MicroRNA expression signatures in solid malignancies. Cancer J. 2012;18:238–43.
Kinose Y, Sawada K, Nakamura K, et al. The role of microRNAs in ovarian cancer. BioMed Res Internat. 2014;2014:11.
Vrba L, Muñoz-Rodríguez JL, Stampfer MR, et al. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS One. 2013;8:e54398.
Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–9R.
Schee K, Fodstad Ø, Flatmark K. MicroRNAs as biomarkers in colorectal cancer. Am J Pathol. 2010;177:1592–9.
Chiosea S, Jelezcova E, Chandran U, et al. Overexpression of dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–50.
Zhang BL, Song FJ, Zheng H, Zhang LN, Zhao YR, Chen KX. SNP rs16917496 within SETS 3′UTR is associated with the age of onset of breast cancer. Chin J Oncol. 2012;23(34):835–7.
Lee AR, Park J, Jung KJ, et al. genetic variation rs7930 in the mir-4273-5p target site is associated with a risk of colorectal cancer. OncoTargets Ther. 2016;9:6885.
Chang S, Sharan SK. BRCA1 and MicroRNAs: emerging networks and potential therapeutic targets. Mol Cells. 2012;34:425–32.
Yang Z, Wu L, Wang AT, et al. dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucl Acids Res. 2016;45(D1):D812–8.
Wang D, Gu J, Wang T, et al. OncomiRDB: a database for the experimentally verified oncogenic and tumor-suppressive microRNAs. Bioinformatics. 2014;30:2237–8.
Petrovic N. miR-21 might be involved in breast cancer promotion and invasion rather than in initial events of breast cancer development. Mol Diagn Ther. 2016;20:97–110.
Petrovic N, Davidovic R, Jovanovic-Cupic S, et al. Changes in miR-221/222 levels in invasive and in situ carcinomas of the breast: differences in association with estrogen receptor and TIMP3 expression levels. Mol Diagn Ther. 2016;20:603–15.
Petrovic N, Kolakovic A, Stankovic A, et al. miR-155 expression level changes might be associated with initial phases of breast cancer pathogenesis and lymph-node metastasis. Cancer Biomark. 2016;16:385–94.
Petrovic N, Mandusic V, Stanojevic B, et al. The difference in miR-21 expression levels between invasive and non-invasive breast cancers emphasizes its role in breast cancer invasion. Med Oncol. 2014;31:867.
Ergun S, Arman K, Temiz E, et al. Expression patterns of miR-221 and its target Caspase-3 in different cancer cell lines. Mol Biol Rep. 2014;41:5877–81.
Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumor Biol. 2015;36:3129–36.
Ergun S, Tayeb TS, Arslan A, et al. The investigation of miR-221-3p and PAK1 gene expressions in breast cancer cell lines. Gene. 2015;555:377–81.
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61.
Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–7.
Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24.
Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mrna-specific upregulation. Int J Genomics. 2014;2014:15.
Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85.
Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucl Acids Res. 2004;32:4776–85.
Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. BBA Mol Cell Res. 2010;1803:1231–43.
Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, exportin 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci USA. 2016;113:E1881–9.
Weber B, Stresemann C, Brueckner B, et al. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–5.
Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14.
Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148:381–92.
Ozsolak F, Poling LL, Wang Z, Liu H, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–83.
Monteys AM, Spengler RM, Wan J, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505.
Huang J-T, Wang J, Srivastava V. MicroRNA machinery genes as novel biomarkers for cancer. Front Oncol. 2014;4:113.
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73.
Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucl Acids Res. 2015;43(D1):D146–52.
Gambari R, Brognara E, Spandidos DA, et al. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: new trends in the development of miRNA therapeutic strategies in oncology (review). Int J Oncol. 2016;49:5–32.
Yu Z, Wang C, Wang M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509–17. doi:10.1083/jcb.200801079.
Xiang J, Wu J. Feud or friend? The role of the miR-17-92 cluster in tumorigenesis. Curr Genom. 2010;11:129–35.
Li Y, Vecchiarelli-Federico LM, Li YJ, Egan SE, Spaner D, Hough MR, Ben-David Y. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood. 2012;119:4486–98.
Habbe N, Koorstra J-BM, Mendell JT, et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–6.
Cha YJ, Lee JH, Han HH, et al. MicroRNA alteration and putative target genes in high-grade prostatic intraepithelial neoplasia and prostate cancer: STAT3 and ZEB1 are upregulated during prostate carcinogenesis. Prostate. 2016;76:937–47.
Li C, Nie H, Wang M, et al. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27:1960–6.
Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33.
Pan X, Wang Z-X, Wang R. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol Ther. 2010;10(12):1224–32.
Almeida AL, Bernardes MV, Feitosa MR, et al. Serological under expression of microRNA-21, microRNA-34a and microRNA-126 in colorectal cancer. Acta Cir Bras. 2016;31(Suppl 1):13–8.
Yang X, Wang X, Shen H, et al. Combination of miR-21 with circulating tumor cells markers improve diagnostic specificity of metastatic breast cancer. Cell Biochem Biophys. 2015;73:87–91.
Dong G, Liang X, Wang D, Gao H, Wang L, et al. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014;31:1–10.
Yu Y, Nangia-Makker P, Farhana L, et al. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98.
du Rieu MC, Torrisani J, Selves J, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56:603.
Sicard F, Gayral M, Lulka H, et al. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013;21:986–94.
Yoruker EE, Terzioglu D, Teksoz S, et al. MicroRNA expression profiles in papillary thyroid carcinoma, benign thyroid nodules and healthy controls. J Cancer. 2016;7:803–9.
Cai G, Qiao S, Chen K. Suppression of miR-221 inhibits glioma cells proliferation and invasion via targeting SEMA3B. Biol Res. 2015;48:37.
Goto Y, Kojima S, Nishikawa R, et al. MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br J Cancer. 2015;113(7):1055–65.
Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–54.
Di Leva G, Gasparini P, Piovan C, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer. 2010;102:706–21.
Stinson S, Lackner MR, Adai AT, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41.
Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117:3155–63.
Petrovic N, Mandusic V, Dimitrijevic B, et al. Higher miR-21 expression in invasive breast carcinomas is associated with positive estrogen and progesterone receptor status in patients from Serbia. Med Oncol. 2014;31:977.
Medimegh I, Omrane I, Privat M, et al. MicroRNAs expression in triple negative vs non triple negative breast cancer in tunisia: interaction with clinical outcome. PLoS One. 2014;9:e111877.
Zhong X, Coukos G, Zhang L. miRNAs in human cancer. In: Fan J-B, editor. Next-generation microRNA expression profiling technology: methods and protocols. Totowa: Humana Press; 2012. p. 295–306.
Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33.
Bhayani MK, Calin GA, Lai SY. Functional relevance of miRNA* sequences in human disease. Mut Res. 2012;731:14–9.
Lee SJ, Seo JW, Chae YS, et al. Genetic polymorphism of miR-196a as a prognostic biomarker for early breast cancer. Anticancer Res. 2014;34:2943–9.
Xia L, Ren Y, Fang X, et al. Prognostic role of common microRNA polymorphisms in cancers: evidence from a meta-analysis. PLoS One. 2014;9:e106799.
Xu Q, Dong Q, He C, et al. A new polymorphism biomarker rs629367 associated with increased risk and poor survival of gastric cancer in chinese by up-regulated miRNA-let-7a expression. PLoS One. 2014;9:e95249.
Xu Y, Liu L, Liu J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer. 2011;128:412–7.
Lee A-R, Park J, Jung KJ, et al. genetic variation rs7930 in the mir-4273-5p target site is associated with a risk of colorectal cancer. Oncol Targets Ther. 2016;9:6885.
Liu Z, Wei S, Ma H, et al. A functional variant at the miR-184 binding site in TNFAIP2 and risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2011;32:1668–74.
Liu H, Gao F, Dahlstrom KR, et al. A variant at a potentially functional microRNA-binding site in BRIP1 was associated with risk of squamous cell carcinoma of the head and neck. Tumor Biol 2016;37:1–10.
Gao F, Xiong X, Pan W, et al. A regulatory MDM4 genetic variant locating in the binding sequence of multiple microRNAs contributes to susceptibility of small cell lung cancer. PLoS One. 2015;10:e0135647.
Cho SH, Ko JJ, Kim JO, et al. 3′-UTR Polymorphisms in the MiRNA machinery genes DROSHA, DICER1, RAN, and XPO5 are associated with colorectal cancer risk in a Korean population. PLoS One. 2015;10:e0131125.
Bruhn O, Drerup K, Kaehler M, et al. Length variants of the ABCB1 3′-UTR and loss of miRNA binding sites: possible consequences in regulation and pharmacotherapy resistance. Pharmacogenomics. 2016;17:327–40.
Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84.
Lembo A, Di Cunto F, Provero P. Shortening of 3′UTRs correlates with poor prognosis in breast and lung cancer. PLoS One. 2012;7:e31129.
He X, Yang J, Zhang Q, et al. Shortening of the 3′untranslated region: an important mechanism leading to overexpression of HMGA2 in serous ovarian cancer. Chin Med J. 2013;127:494–9.
Akman HB, Oyken M, Tuncer T, et al. 3′UTR Shortening and EGF signaling: implications for breast cancer. Hum Mol Genet. 2015;24:6910–20.
Chang J-W, Zhang W, Yeh H-S, et al. mRNA 3 [prime]-UTR shortening is a molecular signature of mTORC1 activation. Nat Commun. 2015;6:7218.
Gruber AR, Martin G, Müller P, et al. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun. 2014;5:5465.
Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500.
He J, Wu J, Xu N, et al. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucl Acids Res. 2012;41:498–508.
Wang Y, Hu C, Cheng J, Chen B, Ke Q, Lv Z, et al. MicroRNA-145 suppresses hepatocellular carcinoma by targeting IRS1 and its downstream Akt signaling. Biochem Biophys Res Commun. 2014;446:1255–60.
Su X, Xing J, Wang Z, et al. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 2013;25:235–9.
Giza DE, Vasilescu C, Calin GA. MicroRNAs and ceRNAs: therapeutic implications of RNA networks. Expert Opin Biol Ther. 2014;14:1285–93.
Yang J, Li T, Gao C, et al. FOXO1 3′UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. Febs Lett. 2014;588:3218–24.
Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8.
Li L, Zhang J, Diao W, et al. MicroRNA-155 and microRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J Immunol. 2014;192:1034–43.
Kan X, Sun Y, Lu J, et al. Coinhibition of miRNA21 and miRNA221 induces apoptosis by enhancing the p53 mediated expression of proapoptotic miRNAs in laryngeal squamous cell carcinoma. Mol Med Rep. 2016;13:4315–20.
Stahlhut C, Slack FJ. Combinatorial action of microRNAs let-7 and miR-34 effectively synergizes with erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle. 2015;14:2171–80.
Bandi N, Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer. 2011;10:55.
Garcia AI, Buisson M, Bertrand P, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279–90.
Chang S, Wang R-H, Akagi K, et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat Med. 2011;17:1275–82.
Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. Febs Lett. 2009;583:3966–73.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Nina Petrovic, Sercan Ergün, and Esma R. Isenovic declare having no conflict of interest.
Funding
This work was supported by the Ministry of Education and Science, Republic of Serbia, Grants OI173049 (B.D.), OI173044 (E.R.I), and Ordu University, Scientific Research Projects Coordination Unit.
Rights and permissions
About this article
Cite this article
Petrovic, N., Ergün, S. & Isenovic, E.R. Levels of MicroRNA Heterogeneity in Cancer Biology. Mol Diagn Ther 21, 511–523 (2017). https://doi.org/10.1007/s40291-017-0285-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-017-0285-9