Abstract
Background
Several targeted therapies have been approved for treatment of solid tumors. Identification of gene mutations that indicate response to these therapies is rapidly progressing. A 34-gene next-generation sequencing (NGS) panel, developed and validated by us, was evaluated to detect additional mutations in community-based cancer specimens initially sent to our reference laboratory for routine molecular testing.
Methods
Consecutive de-identified clinical specimens (n = 121) from melanoma cases (n = 31), lung cancer cases (n = 27), colorectal cancer cases (n = 33), and breast cancer cases (n = 30) were profiled by NGS, and the results were compared with routine molecular testing.
Results
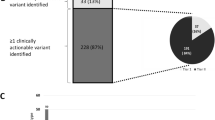
Upon initial mutation testing, 20 % (24/121) were positive. NGS detected ≥1 additional mutation not identified by routine testing in 74 % of specimens (90/121). Of the specimens with additional mutations, 16 harbored mutations in National Comprehensive Cancer Network guideline genes. These various additional mutations were in gene regions not routinely covered, in genes not routinely tested, and/or present at low allele frequencies. Moreover, NGS yielded no false negatives. Overall, NGS detected mutations in 59 % of the genes (20/34) included in the panel, 75 % of which (15/20) were detected in multiple tumor types. Mutations in TP53 were found in 51 % of tumors tested (62/121). Mutations in at least one other (non-TP53) gene present in the panel were detected in 64 % of cases (77/121).
Conclusion
This assay provides improved breadth and sensitivity for profiling clinically relevant genes in these prevalent solid tumor types.



Similar content being viewed by others
References
Arrowsmith J. A decade of change. Nat Rev Drug Discov. 2012;11(1):17–8.
Bailey AM, Mao Y, Zeng J, Holla V, Johnson A, Brusco L, et al. Implementation of biomarker-driven cancer therapy: existing tools and remaining. Gaps Discov Med. 2014;17(92):101–14.
Collins I, Workman P. New approaches to molecular cancer therapeutics. Nature Chem Biol. 2006;2(12):689–700.
Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500.
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39.
Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–16.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–46.
Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene. 2000;19(56):6550–65.
Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, et al. KIT as a therapeutic target in metastatic melanoma. JAMA. 2011;305(22):2327–34.
National Comprehensive Cancer Network. NCCN breast cancer guidelines (version 2.2015). http://www.nccn.org/professionals/physician_gls/pdf/breast.pdf. Accessed 7 April 2015.
National Comprehensive Cancer Network. NCCN colon cancer guidelines (version 2.2015). http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf. Accessed 7 April 2015.
National Comprehensive Cancer Network. NCCN melanoma guidelines (version 3.2015). http://www.nccn.org/professionals/physician_gls/pdf/melanoma.pdf. Accessed 7 April 2015.
National Comprehensive Cancer Network. NCCN non-small cell lung cancer guidelines (version 5.2015). http://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed 7 April 2015.
National Comprehensive Cancer Network. NCCN thyroid carcinoma guidelines (version 2.2014). http://www.nccn.org/professionals/physician_gls/pdf/thyroid.pdf. Accessed 7 April 2015.
National Comprehensive Cancer Network. NCCN soft tissue sarcoma guidelines (version 1.2015) http://www.nccn.org/professionals/physician_gls/pdf/sarcoma.pdf. Accessed 7 April 2015.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–85.
Burrella RA, Swantona C. Tumor heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8(6):1095–1111.
Peng B, Lloyd P, Schran H. Clinical pharmacokinetics of imatinib. Clin Pharmacokinet. 2005;44(9):879–94.
Guo J, Si L, Kong Y, Flaherty KT, Xu X, Zhu Y, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-kit mutation or amplification. J Clin Oncol. 2011;29(21):2904–9.
Venook AP, Arcila ME, Benson AB III, Berry DA, Camidge DR, Carlson RW, et al. NCCN Working Group report: designing clinical trials in the era of multiple biomarkers and targeted therapies. J Natl Compr Canc Net. 2014;12(11):1629–49.
Sánchez-Torres JM, Viteri S, Molina MA, Rosell R. BRAF mutant non-small cell lung cancer and treatment with BRAF inhibitors. Transl Lung Cancer Res. 2013;2(3):244–50.
Kelloff GJ, Sigman CC. Cancer biomarkers: selecting the right drug for the right patient. Nat Rev Drug Discov. 2012;11:201–14.
Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–7.
Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4(1):61–8.
Narita Y, Okamoto K, Kawada MI, Takase K, Minoshima Y, Kodama K, et al. Novel ATP-competitive MEK inhibitor E6201 is effective against vemurafenib-resistant melanoma harboring the MEK1–C121S mutation in a preclinical model. Mol Cancer Ther. 2014;13(4):823–32.
Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29(22):3085–96.
Roock WD, Vriendt VD, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12(6):594–603.
Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza C, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma—rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012;11(2):485–91.
Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–84.
Wang L, Hu H, Pan Y. PIK3CA mutations frequently coexist with EGFR/KRAS mutations in non-small cell lung cancer and suggest poor prognosis in EGFR/KRAS wild type subgroup. PLoS One. 2014;9(2):e88291.
Jänne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14(1):38–47.
Ludovini V, Bianconi F, Pistola L, Chiari R, Minotti V, Colella R, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thoracic Oncol. 2011;6(4):707–15.
Boland JM, Jang J, Li S, Lee J, Wampfler AM, Erickson-Johnson JA, et al. MET and EGFR mutations identified in ALK-rearranged pulmonary adenocarcinoma: molecular analysis of 25 ALK-positive cases. J Thoracic Oncol. 2013;8(5):574–81.
Forbes SA, Beare D, Gunasekeran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database Issue):D805–11.
Lovly C, L Horn, W Pao. Molecular profiling of lung cancer. My Cancer Genome. 2015. http://www.mycancergenome.org/content/disease/lung-cancer. Accessed 7 April 2015.
Balko JM, Mayer I, Levy M, Arteaga C. Molecular profiling of breast cancer. My Cancer Genome. 2015. http://www.mycancergenome.org/content/disease/breast-cancer. Accessed 7 April 2015.
Chan E. Molecular profiling of colorectal cancer. My Cancer Genome. 2015. http://www.mycancergenome.org/content/disease/colorectal-cancer. Accessed 7 April 2015.
Maki RG, Keedy V. Molecular profiling of gastrointestinal stromal tumor (GIST). My Cancer Genome. 2014. http://www.mycancergenome.org/content/disease/gist. Accessed 7 April 2015.
Lovly C, Pao W, Sosman J. Molecular profiling of melanoma. My Cancer Genome. 2014. http://www.mycancergenome.org/content/disease/melanoma. Accessed 7 April 2015.
Espinosa AV, Gilbert J. Molecular profiling of thyroid cancer. My Cancer Genome. 2015. http://www.mycancergenome.org/content/disease/thyroid-cancer. Accessed 7 April 2015.
US National Institutes of Health. https://ClinicalTrials.gov. Accessed 7 April 2015 through 21 December 2015.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signa. 2013;6(269):pl1.
Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–6.
Singh RR, Patel KP, Routbort MJ, Reddy NG, Barkoh BA, Handal B, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15(5):607–22.
Kanagal-Shamanna R, Portier BP, Singh RR, Routbort MJ, Aldape KD, Handal BA, et al. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol. 2014;27(2):314–27.
Qu K, Pan Q, Zhang X, Rodriguez L, Zhang K, Li HR, et al. Detection of BRAF V600 mutations in metastatic melanoma comparison of the Cobas 4800 and Sanger sequencing assays. J Mol Diagn. 2013;15(6):790–5.
Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, et al. Resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18(5):1472–82.
Anaia S, Suzukia K, Ijichia K, Haradaa T, Toyokawac G, Setoc T, et al. A case of crizotinib-resistant lung adenocarcinoma harboring a KRAS mutation and an EML4–ALK fusion gene. J Med Cases. 2014;5(12):631–3.
Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402.
Baselga J, Verma S, Ro J, Huober J, Guardino E, Fang L, et al. Relationship between tumor biomarkers (BM) and efficacy in EMILIA, a phase III study of trastuzumab emtansine (T-DM1) in HER2-positive metastatic breast cancer (MBC). J Cancer Res. 2013;73:Abstract LB-63.
Duzkale H, Shen J, McLaughlin H, Alfares A, Kelly MA, Pugh TJ, et al. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84(5):453–63.
Bai Y, Kim JY, Watters JM, Fang B, Kinose F, Song L, et al. Adaptive responses to dasatinib-treated lung squamous cell cancer cells harboring DDR2 mutations. Cancer Res. 2014;74(24):7217–28.
Chan, E. KRAS c.436G>A (A146T) mutation in colorectal cancer. My Cancer Genome. 2014. http://www.mycancergenome.org/content/disease/colorectal-cancer/kras/28. Accessed 15 Dec 2015.
Akagi K, Uchibori R, Yamaguchi K, et al. Characterization of a novel oncogenic K-ras mutation in colon cancer. Biochem Biophys Res Commun. 2007;352:728–32.
Cescon DW, Bedard PL. PIK3CA genotype and treatment decisions in human epidermal growth factor receptor 2–positive breast cancer. J Clin Oncol. 2015;33(12):1318–21.
Kancha RK, von Bubnoff N, Bartosch N, Peschel C, Engh RA, Duyster J. Differential sensitivity of ERBB2 kinase domain mutations towards lapatinib. PLoS One. 2011;6(10):e26760.
Saito T, Mitomi H, Imamhasan A, Hayashi T, Kurisaki-Arakawa A, Mitani K, Takahashi M, Kajiyama Y, Yao T. PTCH1 mutation is a frequent event in oesophageal basaloid squamous cell carcinoma. Mutagenesis. 2015;30(2):297–301.
Acknowledgments
The authors thank Jeff Radcliff (Quest Diagnostics) for critical review and suggestions on the manuscript, Melissa Smith (MedIncite) for assistance with manuscript preparation, Anne Lin (Quest Diagnostics) for specimen management, Vicki Hartman and Susan-Maria Miranda for coordination of clinical specimens, and Diedre Nguyen for acquisition of clinical result data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Heather Sanders, Kevin Qu, Hairong Li, Lin Ma, Cindy Barlan, Xi Zhang, James Prentice, David Wolfson, Beryl Crossley, Anthony Sferruzza, Feras Hantash, and Frederic Waldman are employees of the Department of Hematology and Oncology, Quest Diagnostics Nichols Institute. John Sninsky, David Ross, Andrew Grupe, and Joseph Catanese are employees of Quest Diagnostics.
Funding
All studies were funded by the Quest Diagnostics Nichols Institute.
Ethical approval and informed consent
The study protocol was reviewed by the Western Institutional Review Board and was determined to be research that is exempt from Institutional Review Board oversight, according to the criteria set forth in 45 CFR 46, “Protection of Human Subjects.” The NGS studies were performed on remnants of specimens collected for routine clinical care that would otherwise have been discarded. Clinical information used in the analyses was recorded in such a manner that subjects could not be identified by investigators, directly or through identifiers linked to the subjects.
Additional information
H. Sanders and K. Qu contributed equally to this work and share first authorship.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sanders, H., Qu, K., Li, H. et al. Mutation Yield of a 34-Gene Solid Tumor Panel in Community-Based Tumor Samples. Mol Diagn Ther 20, 241–253 (2016). https://doi.org/10.1007/s40291-016-0197-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-016-0197-0