Abstract
Background and Objectives
Knowledge obtained via high-throughput technologies, used for tumor genome sequencing or identifying gene expression and methylation signatures, is clinically applicable thanks to molecular characterization in the context of tumor development and progression. This study was conducted to assess the impact of specific KRAS mutation in codons 12 and 13 on clinical outcome of chemotherapy and radiotherapy in colorectal cancer patients.
Methods
A total of 239 samples of colorectal adenocarcinoma underwent histological evaluation and DNA isolation.
Results and Conclusions
Patients with a mutation in KRAS codon 13 experienced worse outcome than those with a mutation in KRAS codon 12. Moreover, the cases of mutations in KRAS codons 12 or 13 were associated with a significantly higher mortality than the cases of wild-type KRAS, and some patients with KRAS mutated in codon 12 had an exceptionally long overall survival. Finally, primary preoperative radiation therapy followed by surgery significantly increased overall survival more efficiently than surgery followed by chemotherapy. This should be investigated in further studies. The fact that all patients treated with radiotherapy + surgery were alive, again focused our attention on the effect of preoperative radiation therapy on the prognosis for colorectal cancer patients. However, the number of patients in this subgroup is too small to allow any specific explanation for this observation. We should, rather, point out a problem for further investigation.
Similar content being viewed by others
We determined the differences in treatment outcome in 100 colorectal cancer patients with wild-type (WT) KRAS and 139 patients with mutations in exon 2. The median overall survival of the patients with KRAS mutations in codons 12 or 13 was shorter versus patients with WT KRAS (19 vs. 29 months) |
We assessed whether tumors with mutations in KRAS codon 12 represent a less aggressive subtype compared with those with mutations in KRAS codon 13. In fact, patients with KRAS mutations in codon 12 had longer median overall survival compared with the KRAS mutated in codon 13 (25 vs. 18 months, respectively) |
Patients with KRAS mutation have better median overall survival when treated with radiotherapy + surgery than surgery + chemotherapy (32 vs. 19 months, respectively) |
1 Introduction
High-throughput technologies such as microarrays or new-generation sequencing techniques enable the determination of the genomic sequences of tumors or the identification of gene expression and methylation signatures. However, the unprecedented use of such knowledge in clinical practice is not enabled by the sole detection of new mutations, but rather by their profound molecular characterization in the context of tumor development and progression. The characterization of mutations in the APC gene that cause familial adenomatous polyposis of the large intestine has led the way for routine oncological prophylaxis. Prophylactic tests have been expanded with mutation analysis in DNA mismatch repair genes, such as MLH1, MSH2, MSH6 or PMS2. In the last decade, gene expression analyses were conducted in order to predict the likelihood of disease recurrence, especially in stage II colon cancer patients treated with surgery and fluorouracil/leucovorin [1]. Finally, for 15 years, attempts have been made to create the CpG island methylator phenotype for colorectal cancer [2].
However, it was the discovered correlation between mutations in the KRAS gene and the efficacy of treatment using a monoclonal antibody targeting epidermal growth factor receptor (EGFR), such as panitumumab and cetuximab, that became a milestone in personalized oncology. Colorectal cancer cells carrying somatic KRAS mutations [3] are less likely to respond to EGFR inhibition. On the other hand, recent studies show that wild-type (WT) KRAS status is not the only factor crucial for patient qualification for targeted therapy with an anti-EGFR monoclonal antibody. Patients bearing BRAF or KRAS mutations (in exon 3 and 4, respectively) had poorer response rate to cetuximab and irinotecan compared with the WT KRAS and BRAF patients [3]. Also, high-throughput technologies (such as massively parallel tumor multigene sequencing) were used to evaluate the response to anti-EGFR monoclonal antibodies. In this study, treatment with panitumumab was associated with longer progression-free survival (PFS) among the WT KRAS (codons 12/13/61), WT NRAS and WT BRAF patients [4].
In this study, we wanted to evaluate whether tumors with mutations in KRAS codon 12 represent a less aggressive subtype compared with those with mutations in KRAS codon 13, and correlate patients with KRAS mutations in codon 12 or 13 versus WT KRAS. Finally, we wanted to access the differences in the treatment outcome in KRAS WT and mutated metastatic colorectal cancer (mCRC) patients.
2 Methodology
2.1 Patient Selection
Overall, 239 patients with an histologically established diagnosis of colorectal adenocarcinoma (after surgery), for whom the collection of tissue samples was part of their routine care and treatment, were included in the study.
The reviewed cohort of WT KRAS (n = 100) and mutant KRAS (n = 139) specimens of colorectal carcinoma was obtained from the Molecular Oncology and Genetics Unit, Department of Tumor Pathology and Pathomorphology, The F. Lukaszczyk Oncology Center, Bydgoszcz, Poland. Informed consent for mutation testing was obtained from all patients.
All patients were treated using surgery (between July 2006 and December 2012) and adjuvant therapy (chemotherapy standard or radiation therapy). Progression of metastatic disease occurred in all patients, at different times after the first surgery. Treatment with cetuximab or panitumumab was carried out between July 2008 and February 2013 (anti-EGFR agent in third-line of chemotherapy was used). Table 1 presents the clinical characteristics of all 239 patients.
In each case, a representative sample of the adenocarcinoma tissue area, a formalin-fixed, paraffin-embedded (FFPE) block or a cytology specimen was identified by a pathomorphologist and scraped for DNA isolation. Histological evaluation and DNA isolation were performed as previously described [5].
2.2 Mutation Analysis
Each individual case was evaluated for KRAS mutations in exon 2 (codons 12 and 13) using one of the following methods of mutation analysis: single-strand conformation polymorphism (SSCP), Sanger sequencing, or CE-IVD polymerase chain reaction (PCR)-related methods: strip test containing allele-specific probes (ViennLab) and real-time PCR methods using Taqman probes (EntroGen, Inc.), high resolution melting-curve analysis (TibMolBiol) and allele-specific amplification ARMS and detection by Scorpions technology (Therascreen). Additional analysis of mutations in KRAS exon 3 or BRAF exon 15 was performed for a number of selected patients diagnosed with mCRC between September 2012 and February 2013, previously evaluated using the real-time PCR methodology (Entrogen, Inc.) or PCR with reverse-hybridization (ViennaLab). Uncertain results were double-checked in a second analysis performed using another molecular method from the list above.
2.3 Statistical Analysis
For statistical analysis, the STATISTICA (version 10.0) computer software (StatSoft, Inc.) was used. Differences between the categorized groups were assessed using the log-rank test. Correlations between overall survival and the results of KRAS mutation analysis in codons 12 and 13 were estimated using the Kaplan–Meier estimator. The results were considered statistically significant at p < 0.05.
3 Results
3.1 Characteristics of the Patients
Clinical characteristics for 239 patients, such as mean age, sex, tumor site, and treatment, are provided in Table 1. The way in which colorectal cancer affects the daily living abilities of the patients was assessed using the Eastern Cooperative Oncology Group (ECOG) performance status (Table 1) [http://ecog.dfci.harvard.edu/general/perf_stat.html]. The majority of mCRC patients (83 %) were outpatients and were able to carry out work of a light or sedentary nature, while the second group (14 %) represented ECOG grade 2, in which patients are capable of all self-care but unable to carry out any work activities. The smallest group (3 %) belonged to patients capable of only limited self-care (ECOG grade 3). No completely disabled patients were included in this study.
3.2 Specimen Evaluation and Molecular Analysis of KRAS Exon 2
A total of 239 FFPE and cytology samples were qualified for molecular analysis using quantitative scale (QS), and the percentage of tumor cells (PTC) by pathomorphologists at the F. Lukaszczyk Oncology Center, as previously described [5, 6]. Among the 239 samples qualified for the determination of the presence of KRAS mutations, 139 (57.4 %) had a somatic mutation in codon 12 or 13. The majority of KRAS mutations were diagnosed in codon 12 (n = 109), compared with 30 specimens with mutations in codon 13. Most of the mutations (95.8 %) were detected with the PTC at 10 % or above. Furthermore, real-time PCR allowed the detection of KRAS p.G12D, p.G12V, p.G12A, or p.G12D in six samples with a low PTC (below 10 %).
3.3 Impact of Specific KRAS Mutation in Exon 2 on Clinical Outcome
This study was conducted to assess whether tumors with mutations in KRAS codon 12 represent a less aggressive subtype compared with those with mutations in KRAS codon 13. A comparative analysis of overall survival in correlation with potential differences was conducted between the patient groups with mutant and WT KRAS.
The median overall survival in patients with KRAS mutations in codon 12 was better compared with patients with KRAS mutations in codon 13 (25 vs. 18 months, respectively) [Fig. 1a]; the results were at the borderline of statistical significance. Unusually long overall survival (above 80 months, up to 160 months) was observed in 11 patients of the group with codon 12 mutations.
Kaplan–Meier curves of KRAS status. a In the subset of patients with mutant KRAS tumors, mutation in codon 12 is not correlated with survival. b Comparing overall survival in patients with mutant KRAS tumors versus wild-type KRAS tumors, mutations in codons 12 or 13 are significantly correlated with better survival. OS overall survival
Various types of KRAS mutations were tested for their potentially different effect on the patients’ overall survival. Median overall survival of patients with KRAS mutations in codons 12 or 13 was 19 months, while that of patients with WT KRAS was 29 months (p = 0.03, log-rank test) [Fig. 1b].
3.4 Long-Term Results Depending on the Method of Treatment
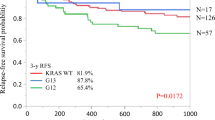
We assessed whether Polish patients with a new diagnosis of colorectal cancer, after surgical removal of the primary tumor with KRAS mutations in codon 12 or 13, might have a better prognosis regarding overall survival than patients with WT KRAS. To learn the effect of the treatment regimen on the remote results, we performed a comparative analysis in the group of patients with KRAS mutated in codon 12 according to the administered therapy (Fig. 2, blue and green lines). As it occurred, the median overall survival of patients carrying the somatic KRAS mutation and treated with radiotherapy + surgery (RT + SUR) was considerably better than that of patients treated with surgery + chemotherapy (SUR + CT) [32 vs. 19 months, respectively].
Finally, we sought to determine the differences in the treatment outcome in 100 colorectal cancer patients with WT KRAS and 139 patients with mutations in codon 12 or 13. It was also revealed that in the subgroup of patients carrying a KRAS mutation in codon 13 and treated with RT + SUR (a small group of seven patients), all patients remained alive (Fig. 2, the dotted purple line). Among patients with KRAS mutated in codon 13 treated with SUR + CT, 70 % remained alive (Fig. 2, red line). However, median overall survival in both subgroups did not differ significantly.
4 Discussion
In the recent years, the recommendations for somatic mutation analysis, conducted as part of the qualification of patients for immunotherapy using monoclonal antibodies, were limited only to the second exon of the KRAS gene. Advances in targeted therapies in oncology, as well as constantly increasing knowledge in the field of colorectal cancer biology, have participated in the expansion of the range of analyses in molecular diagnostics with KRAS exons 3 and 4 and new genes, such as NRAS or BRAF (Fig. 3). On the other hand, before this expansion of the panel of analyzed somatic mutations, some patients carrying mutations in KRAS, BRAF, or NRAS had initiated treatment with cetuximab or panitumumab [3, 5], which resulted in the development of primary resistance. However, patients with WT BRAF/NRAS/PI3KCA and KRAS tumors have the highest chance of response to the epidermal growth factor receptor-targeted therapy [7–9]. It is known that KRAS mutations in codon 12 are associated with a mucinous phenotype of colorectal cancer. A significant characteristic of the colorectal cancers associated with KRAS codon 13 mutations is that they are rather non-mucinous, but characterized as more aggressive tumors with a greater metastatic potential [9, 10]. In fact, we observed a 7-month difference in overall survival in favor of patients with a KRAS mutation in codon 12. A more precise analysis of this group might determine a subtype of mutation with a particularly good prognosis. The results of De Roock et al. from pooled analysis from seven clinical trials [11], and detailed retrospective analysis of colorectal patients with KRAS codon p.G13D mutation who received cetuximab [12], indicate that there is no black and white dogma that only mCRC patients with WT KRAS benefit from cetuximab [13]. Interestingly, similar results to ours, in which patients with a mutation in KRAS codon 13 experienced worse outcome compared with the KRAS codon 12 mutants, were observed in lung cancer [14]. Second, we demonstrated that the cases of mutations in KRAS codons 12 or 13 were associated with a significantly higher mortality compared with the cases of WT KRAS (median overall survival: 19 vs. 29 months, respectively; Fig. 1b). In the group of patients with mutations in KRAS codon 12 participating in our study, exceptionally long overall survival (up to 160 months) was observed in 11 patients. This suggests that in the group with KRAS mutated in codon 12 there is a subgroup of several patients with a particularly good prognosis. The most comprehensive explanation for this observation would be provided by thorough genotyping of KRAS codon 12 and BRAF/NRAS/PI3KCA somatic mutation screening to determinate mutation status that correlates with the good prognosis. For example, mutations c.35G>A (p.G12D) and c.35G>C (p.G12A) [Fig. 1a] confer less potent transforming ability than c.34G>C (p.G12R) and c.35G>T (p.G12V) [15]. Our observations emphasize the importance of the fact that various mutations (even in a single exon) may contribute to different characteristics of the tumor, supporting its unique morphology [12, 16]. A growing number of reports underline the prognostic and predictive value of the information derived not only from the analysis of somatic mutations, but also from the analysis of miRNA expression [17].
Molecular diagnostics in colorectal cancer: examples of mutation analysis in the KRAS, BRAF, and NRAS genes. a Analysis of KRAS mutations in codons 12 and 13 using the real-time PCR methodology with melting curve analysis. No KRAS mutations detected in any of the two codons (the presence of the following mutations was tested: 12D, 12C, 12V, 12A, 12R, 12S, 12T, 13D, 12D/13D and 13C). Each curve represents the time course of the real-time PCR assay. Line 1 represents the detection of a control WT amplicon, line 2 represents control 13C, line 3 represents control 12C, and line 4 represents sample of interest with WT KRAS status. b Analysis of the BRAF V600E and KRAS codons 12 and 13 mutations using PCR with reverse hybridization. Blue line positive signal detected with a control PCR reaction, no mutation detected in KRAS codons 12 or 13 or BRAF V600E. The tested mutations are listed on the right. c, d Analysis of NRAS mutations in exons 2, 3, and 4 using allele-specific primers. c Filter: FAM 465-510; amplification curves represent the following PCs in exon 2: G12C, G12D, G12S, G13V, G13R; in exon 3: Q61K, Q61R, Q61L, Q61H; and in exon 4: A146T (C p in the range 27.03–29.03). Baselines represents no amplification with primers complementary to mutant NRAS variants in exons 2, 3, and 4, and NTC. d IC for VIC/yellow reporter measuring DNA load of the tested sample. Amplification curves represent internal controls for each of the 10 reactions (C p in the range 29.23–29.54), flat lines represent NTC. PCR polymerase chain reaction, WT wild-type, PCs positive controls, IC internal controls
Surprising results from this study were obtained by comparing overall survival of patients divided into subgroups according to the KRAS mutations in codons 12 or 13 and the administered therapy. All patients were subjected to surgical resection (SUR) of colorectal cancer combined with adjuvant therapy in the form of standard chemotherapy (oxaliplatin) [CHT] or primary preoperative radiation therapy (RH) followed by surgery (SUR). It seems that tumor responsiveness to therapy (radiation versus chemotherapy) may be predicted by DNA content (mutation in KRAS codon 13). On the other hand, it has been recently showed on a similar number of CRC patients that tumor ploidy is not associated with tumor response to radiation [18]. Therefore, more detailed molecular analysis (somatic mutations, microsatellite instability, chromosomal instability and genome-wide association studies) on a bigger cohort of CRC patients should be performed to evaluate and better understand the response to radiotherapy.
5 Conclusions
We showed worse median overall survival of patients with KRAS mutations in exon 2 than KRAS WT. In particular, patients with mutation in codon 13 and treated without radiotherapy had the lowest overall survival from all studied groups. Also, the fact that all patients treated with radiotherapy and surgery were alive, again focused our attention on the effect of preoperative radiation therapy on the prognosis for colorectal cancer patients. However, the number of patients in this subgroup was too small to allow any specific explanation of this observation. We should, rather, point out a problem for further investigation.
References
O’Connell MJ, Lavery I, Yothers G, Paik S, Clark-Langone KM, Lopatin M, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28(25):3937–44.
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–6.
Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101(4):715–21.
Peeters M, Oliner KS, Parker A, Siena S, Van Cutsem E, Huang J, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res. 2013;19(7):1902–12.
Lewandowska MA, Jozwicki W, Zurawski B. KRAS and BRAF mutation analysis in colorectal adenocarcinoma specimens with a low percentage of tumor cells. Mol Diagn Ther. 2013;17(3):193–203.
Lewandowska MA, Jozwicki W, Jochymski C, Kowalewski J. Application of PCR methods to evaluate EGFR, KRAS and BRAF mutations in a small number of tumor cells in cytological material from lung cancer patients. Oncol Rep. 2013;30(3):1045–52.
Pietrantonio F, Perrone F, Biondani P, Maggi C, Lampis A, Bertan C, et al. Single agent panitumumab in KRAS wild-type metastatic colorectal cancer patients following cetuximab-based regimens: clinical outcome and biomarkers of efficacy. Cancer Biol Ther. 2013;14(12):1098–103.
Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023–34.
Custodio A, Feliu J. Prognostic and predictive biomarkers for epidermal growth factor receptor-targeted therapy in colorectal cancer: beyond KRAS mutations. Crit Rev Oncol/Hematol. 2013;85(1):45–81.
Bazan V, Agnese V, Corsale S, Calo V, Valerio MR, Latteri MA, et al. Specific TP53 and/or Ki-ras mutations as independent predictors of clinical outcome in sporadic colorectal adenocarcinomas: results of a 5-year Gruppo Oncologico dell’Italia Meridionale (GOIM) prospective study. Ann Oncol. 2005;16 Suppl 4:iv50–5.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62.
De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p. G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304(16):1812–20.
Kirk R. Genetics: in colorectal cancer, not all KRAS mutations are created equal. Nat Rev Clin Oncol. 2011;8(1):1.
Metro G, Chiari R, Duranti S, Siggillino A, Fischer MJ, Giannarelli D, et al. Impact of specific mutant KRAS on clinical outcome of EGFR-TKI-treated advanced non-small cell lung cancer patients with an EGFR wild type genotype. Lung Cancer. 2012;78(1):81–6.
Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312(5989):71–5.
Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10(1):13–27.
Cho WC. Epigenetic alteration of microRNAs in feces of colorectal cancer and its clinical significance. Expert Rev Mol Diagn. 2011;11(7):691–4.
Fadhil W, Kindle K, Jackson D, Zaitoun A, Lane N, Robins A, et al. DNA content analysis of colorectal cancer defines a distinct ‘microsatellite and chromosome stable’ group but does not predict response to radiotherapy. Int J Exp Pathol. 2014;95(1):16–23.
Acknowledgment and Disclosures
We would like to thank Dr. Z. Pawlowicz for generating the conditions advantageous for our research. Krzysztof Roszkowski, Bogdan Zurawski, Wojciech Jozwicki, Pawel Basta, and Marzena Anna Lewandowska report no financial disclosures. This work is supported by a Foundation for Polish Science, cofinanced from European Union, Regional Development Fund ( Homing Plus/2010-2/7).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Roszkowski, K., Zurawski, B., Jozwicki, W. et al. Impact of Specific KRAS Mutation in Exon 2 on Clinical Outcome of Chemotherapy- and Radiotherapy-Treated Colorectal Adenocarcinoma Patients. Mol Diagn Ther 18, 559–566 (2014). https://doi.org/10.1007/s40291-014-0107-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-014-0107-2