Abstract
There is emerging interest in the ability of the intestinal microbiome—the collective genome of resident microbes—to impact host function. Given the dynamics of development, the pediatric microbiome is of particular importance as alteration of microbial communities (microbiota) early in life may deeply influence long-term health and disease. This review explores the current understanding of the establishment and evolution of the microbiome in early life. It then discusses the evidence for three key spheres of microbiome-mediated disease in pediatrics: inflammatory bowel disease, obesity and atopic disease. The role of microbiota-derived therapeutics is also examined through the lens of future care for children. Probiotics and fecal microbiota transplant, a novel therapy to restore the microbiome, are explored and the rationale for caution outlined for infants and children. Ultimately, there is potential for research born out of the field to alter our paradigm of pediatric disease and its treatment.
Similar content being viewed by others
Introduction
With the recent emergence of culture-independent sequencing techniques we are just beginning to understand the rich and diverse microbial landscape in both children and adults. The gastrointestinal (GI) system alone contains over 100 trillion microbes with complex diversity and heterogeneity across individuals [1, 2]. The perpetual interplay between microbial communities (microbiota), and human hosts has an impact on health and susceptibility to disease.
Although the impact of the microbiota in GI disease has been robust, there is growing interest in the ability of the intestinal microbiota to influence host functions beyond the GI system. The microbiome appears to have a broad reaching impact on human health via modulation of immune homeostasis, metabolism, colonic colonization resistance and possibly alteration of neuro-active metabolic pathways that affect behavior [3, 4].
Given the dynamics of the microbiota in early life, it is speculated that infants and children are particularly sensitive to alternations in the microbiota and the associated downstream health consequences, suggesting a ‘developmental window’ in establishing a healthy microbiome.
Interestingly, host genetics may play a role in the establishment of the microbiome, as specific genetic loci appear to influence bacterial microbiome composition [5, 6]. In this vein, the ‘early-life programming hypothesis’ proposes that the energy metabolism of early life microbiota and the interaction of these microbes with the immune system help determine the risk for diseases later in life, possibly mediated by epigenetics [7]. Additionally, early commensal microbiota is also important in the establishment of a balanced helper T cell (Th1/Th2) immune response to antigenic stimuli more directly [8]. Accordingly, pediatricians and scientists must become familiar with this rapidly evolving field.
The prospect of harnessing the translational potential of this field has engaged funding agencies to invest in human microbiome consortiums. In 2007, the United States National Institutes of Health initiated the 171 million dollar Human Microbiome Project (HMP), and in 2008, the European Commission funded the 22 million euro Metagenomics of the Human Intestinal Tract (Meta-HIT) Consortium [3, 9, 10]. The HMP and Meta-HIT Consortium serve as beacons for the promise of a new era of personalized medicine.
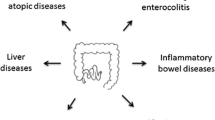
Given the emerging importance of the microbiome in infants and children, the following review focuses on the current state of evidence on the establishment and evolution of the human microbiome, highlights the nexus of dysbiosis—pathologically deviated microbiota—in three representative pediatric disease examples, and provides insight into microbiota-derived therapeutics for infants and children.
Lessons from the Adult Microbiome Literature
The exploration of the microbiome in adults preceded evaluation of the pediatric population, and accordingly lessons can be learned about the direction of future research in children.
The progress of DNA-based, culture-independent techniques used to assess the microbiota in adults has primed pediatric-oriented researchers with the optimal tools to explore the microbiome in infants and children (Table 1) [11••]. The detail of the varied techniques are beyond the scope of this paper and well described elsewhere [11••]. Briefly, traditional in vitro culture methods used to grow bacteria cultivate less than 30 % of bacterial flora in the human GI system. Accordingly, researchers typically extract microbial DNA from stool and subject it to high throughput, next generation sequencing of the variable regions of the bacterial 16S rRNA gene [11••] (Table 1). The relationship between 16S rRNA gene sequences and comparison to a reference database of known bacteria allows for the phylogenic classification of the bacterial community.
The definition of a ‘normal’ microbiome is complex in adults, as the commensal flora exhibit redundancy and diversity that continue to evolve as next generation sequencing better characterizes the microbial community. The complex field and its challenges are explored more thoroughly elsewhere [2, 12, 13]. Briefly, core functional clusters of bacteria, termed enterotypes, in adults are characterized by prominent microbial genera: Bacteroides, Prevotella and Ruminococcus, respectively. Assessing the proportions of the dominant genera that comprises a healthy microbiota is key in determining if alterations are associated with disease; however, distinct enterotypes have yet to be clearly established in children and may differ depending on a child’s age, in turn, making the definition of 'normal' GI microbiota in children challenging [10, 14]. The concept of dominant enterotypes, changes in microbial diversity, and a systems biology shift toward bacterial and possibly virome groups leading to pathology, rather than a single microbial agent as the cause of pathology, is emerging as the prevailing perspective [15]. Specifically, there is evidence to suggest that patients with recurrent Clostridium difficile infection (CDI) associated-diarrhea have a significant decrease in fecal microbiome diversity, whereas patients with celiac disease have higher diversity compared with controls [15–17]. Similarly, patients with inflammatory bowel disease (IBD) appear to have community microbial dysbiosis as compared to the presence of a single ‘causative’ organism, although speculation persists regarding the reduced abundance of Faecalibacterium prausnitzii in Crohn’s disease and Akkermansia muciniphila in ulcerative colitis compared to controls [6, 15, 18]. In adults, irritable bowel syndrome, rheumatoid arthritis, obesity, metabolic syndrome, atopic disease, and neuro-psychiatric illness appear to have meaningful differences in gut microbiota compared with controls; however, the pediatric microbial composition in these diseases has not been fully explored. Importantly, emerging animal models and human fecal microbiota studies are beginning to suggest the microbiome as part of the cause, as opposed to consequence of disease [4, 11••, 15, 18–20].
Establishment and Evolution of Human Microbiome
Recent molecular studies of meconium from small numbers of newborns suggest the fetal gut may not be sterile as previously thought, although the mechanism of colonization remains unclear [21, 22]. The composition of the newborn microbiome is determined in part by the birthing process when the baby is exposed to maternal microbiota [15]. As anticipated, babies born vaginally are colonized with maternal microbiota from the birth canal such as Lactobacillus sp., Sneathia sp. and Prevotella sp., whereas babies born by Cesarean section are colonized with common skin microbes such as Staphylococcus sp., Propionibacterium spp. and Corynebacterium sp. [15, 23]. During the first few months of life, GI bacterial diversity generally is low compared with adulthood. However, this changes rapidly and diversity dramatically increases by 3 months of life [15, 24]. By 11 months of life the initial colonizers are supplanted by individually unique infant-specific phylotypes, which are distinct from their mother [15, 25]. An adult-oriented microbiota begins to emerge at the end of the first year of life and is fully entrenched by 2.5 years of age, largely stable until old age [15].
In addition to the birthing delivery modality, many factors contribute to the composition of the pediatric microbiome, including maternal weight and early diet [15, 26]. Breast milk is not sterile, containing a variety of bacteria, and breast-fed infants are more likely to be initially colonized with Bifidobacteria sp. compared with formula-fed infants [15, 26, 27]. Breast milk also contains oligosaccharides that promote colonization with commensal organisms and antimicrobial factors such as lysozyme, lactoferrin and antibodies that prevent colonization with potential pathogens [7, 26, 27]. Other important factors that influence the evolution of the microbiota during early life include prolonged exposure to the healthcare setting (e.g. prematurity), antibiotic exposure (frequency, duration, type), the timing and type of solid food introduction, geographic location, and the local environment (e.g. farming, urban) [15, 27, 28]. For example, although methodologically limited, the introduction of solid food was associated with an increase in Bacteroidetes in an infant and whole genome analysis revealed an increase in bacterial genes associated with carbohydrate digestion compared with the time period prior to weaning [29]. Overall, the pediatric microbiome appears to undergo rapid early development and is sensitive to many external factors before long-term stability is established.
Microbial Dysbiosis and Pediatric Disease
Inflammatory Bowel Disease (IBD)
The intestinal microbiota is thought to play a role in the pathogenesis of IBD—a chronic inflammatory disorder of the GI tract—as opposed to the consequence of the disease [30–32]. Although the precise mechanism in which the microbiome interacts with host genetic, environmental factors, and the immune system is still under exploration, animal studies and clinical observations are promising. For example, mutations in NOD2 and ATG16L1, IBD risk alleles, are associated with a relative abundance of members of the Faecalibacterium and Escherichia genera compared to controls [33, 34]. Additionally, a large metagenomic sequencing study has highlighted a statistically significant difference comparing the bacterial microbiota in Crohn’s disease, ulcerative colitis and healthy controls [35]. This concept was strengthened in the pediatrics literature with a recent prospective, multi-center study that highlighted the role of the microbiome in children with IBD [36]. The authors noted significantly less microbial species diversity in the gut microbiome of children with ulcerative colitis compared to controls. Most notably, steroid non-responders harbored less bacterial diversity compared with steroid-responsive children, signaling a potential prognostic marker [36]. Given the influence of the microbiome, there remains speculation regarding the role of antimicrobial agents as a risk factor for the development of IBD in the pediatric population. A recent pediatric population-based cohort study has helped shed light on this association [37•]. Kronman et al. [37•] examined over one million children and noted an 84 % relative risk (RR) increase for IBD in children prescribed antibiotics such as clindamycin, that target anaerobic bacteria present in the gut (unexposed 0.83/10,000 person-years; exposed 1.52/10,000 person-years). A dose–response relationship between antibiotic use and IBD was found as each antibiotic course increased the IBD hazard by 6 %. Antibiotic therapy early in life (before 1 year of age) was associated with the highest adjusted hazard ratio for IBD (5.51, 95 % confidence interval, CI 1.66–18.28) but decreased significantly over the age span [37•]. Although, these findings are observational, it is another example of the importance of appropriate antibiotic stewardship, particularly early in infancy.
Obesity
The role of the intestinal microbiota is believed to extend beyond the GI tract. There is mounting evidence regarding the impact of microbes on obesity by modulation of host metabolism [19, 38, 39]. Most recently, Ridaura et al. [40••] demonstrated the impact of the microbiome in obesity in an elegant human–mouse model. Fecal microbiota were transplanted from adult human female twin pairs that were discordant for obesity into germ-free mice. Mice receiving the microbiota from the lean twin remained lean, while mice receiving microbiota from the obese twin had increased weight gain. Furthermore, cohousing the obese microbiota mice with lean microbiota mice prevented the development of obesity and the associated metabolic phenotype in the latter mice, as they acquired the lean microbiota including increased numbers of the phylum Bacteroidetes [40••].
Additional evidence supports a role for the gut microbiota in determining animal weight and metabolism. Traditionally raised mice have significantly more body fat compared to germ-free mice, and germ-free mice colonized with normal flora develop hepatic lipogenesis and decreased glucose tolerance [41, 42].
Although speculation remains on the precise mechanism, some have implicated the phylums of Bacteroidetes and Firmicutes in the pathogenesis of obesity, because obese mice have fewer Bacteroidetes and increased numbers of Firmicutes in their GI tract. Experimental evidence suggests that this predisposes animals to energy storage and obesity [18, 43]. A small (n = 49) Finnish study utilizing fluorescent in situ hybridization to characterize the fecal microbiome showed infants with lower Bifidobacterium and higher Staphylococcus aureus in their stool had an increased risk of obesity based on the International Obesity Task Force criteria [44]. A larger clinical study (n = 175) of Kazakh school children found a negative correlation between body mass index and Bacteroidetes number and Bacteroidetes/Firmicutes ratios, supporting the animal model evidence [45]. Interestingly, this association was only observed in females and the authors speculated that this was related to differential iron metabolism among genders that impacts the composition of the intestinal microbiota [45]. Although there is a need for ongoing research, the prospect of microbiota-driven therapeutics to help ameliorate the burden of childhood obesity is welcomed by all stakeholders.
Atopic Diseases
Several studies have looked at the relationship between the early microbiome and subsequent development of allergic disease with conflicting results. A recent study of 98 infants at risk for allergic disease found that decreased microbial diversity from fecal samples taken at 1 week of life was associated with increased risk for eczema, but not atopy, at 1 year of age compared with infants with a more diverse gut microbiota [46]. In contrast, a cohort study of 411 children born to mothers with asthma found that low microbial diversity at 1 month and at 1 year of age was associated with childhood atopy during the first 6 years as measured by immunoglobulin E (IgE) levels, skin testing, and eosinophilia [47]. Interestingly, no association was found for asthma or eczema with microbial diversity [47]. A third study of 47 infants with fecal samples for microbial diversity at 1 month and 1 year of age found an association between low diversity and asthma at 7 years of age but not other allergic diseases [48]. This study is limited by the small sample size with only eight children developing asthma. Collectively, these results demonstrate that the time-point chosen to examine both the diversity of the evolving microbiota and the clinical outcome in the developing child can profoundly impact the study results.
The hygiene hypothesis postulates that reduced exposure to microbes, including those in the GI tract, during the early programming period leads to an exaggerated Th2 allergic response [8]. This suggests that reductions or alterations in early microbiota from antibiotic utilization in infancy may predispose individuals to Th2 mediated atopic disease later in life. Again, there is conflicting evidence with prospective studies both in favor and against a role for early antibiotic use and risk for atopic disease [8]. However, a trend is emerging favoring an association between early antibiotic use, including during pregnancy, and allergy and asthma. A recent study of two cohorts, including over 30,000 children from the Danish National Birth Cohort followed for 5 years, found an increased risk for asthma hospitalization (RR 1.17, 95 % CI 1.00–1.36) and the use of oral corticosteroids (RR 1.18, 95 % CI 1.10–1.27) for children born to mothers who took antibiotics during pregnancy [49]. A second longitudinal cohort study of 4,952 children found a striking dose-dependent response associating antibiotic use (before 2 years of age) with asthma, eczema and hay fever at age 7 years [50••]. The authors highlighted the odds ratio (OR) of having asthma with four or more courses of antibiotics was 2.82 (95 % CI 2.19–3.63) compared with a single antibiotic course (OR 1.11, 95 % CI 0.84–1.48) [50••].
Whether these associations are solely due to alterations in gut microbiota is unclear. Certain antibiotic classes such as the macrolides have immune-modulatory effects and may inherently promote a Th2 response independent of any direct bacterial killing [8]. Overall, the nature of the relationship between antibiotic therapy and atopy remains unresolved, and continues as an area of ongoing research.
Emerging Therapeutic Implications
Probiotics
Probiotics have been defined by the World Health Organization [51] as 'live microorganisms which, when administered in adequate amounts, confer a health benefit to the host'. A number of different bacteria are currently being used as probiotics, most commonly Lactobacillus and Bifidobacteria sp. Multiple studies have examined whether probiotic administration can prevent or treat atopic, inflammatory, and metabolic diseases. However, issues central to heterogeneity of probiotics have hampered precise evaluation in this field. Given immense variations in probiotics, doses, lengths and populations, there have been challenges in comparing outcomes across studies. Additionally, if one speculates on the association between microbial diversity in the gut and disease, 98 children given L. rhamnosus GG had no change in microbial diversity compared to controls, highlighting the uncertainty of a single microorganism shifting overall microbial diversity [52]. With these strong cautions in place, and despite heterogeneity in this research sphere, evidence is emerging that probiotics early in life may impact disease in later childhood.
Two recent meta-analyses concluded that early probiotic administration during pregnancy and/or in early childhood provides some protection against atopic disease [53, 54]. Elazab and colleagues identified 25 studies involving 4,031 children for analysis. Measured outcomes included total IgE levels, atopic sensitization as determined by skin prick test, and the presence of asthma or wheezing [53]. Early probiotics administered either during pregnancy or postnatally resulted in decreased IgE levels. There was also an association between probiotic administration and reduced atopy but only when the probiotics were given both pre- and postnatally (Table 2) [53]. No association was found between probiotics and asthma/wheezing. L. rhamnosus GG is an American Type Culture Collection strain of L. rhamnosus that commonly used for probiotic studies and was the most represented probiotic in studies included in this meta-analysis. However, a wide variety of probiotics were used and L. acidophilus was associated with increased atopic sensitization (Table 2) [53].
A second meta-analysis by Pelucchi et al. [54] examined the relationship between probiotic supplementation pre- and postnatally and atopic dermatitis (Table 2). The authors examined 18 publications from 14 study populations and concluded that probiotics offer moderate protection against atopic dermatitis regardless of whether the probiotics are administered prenatally, postnatally or both (Table 2) [54]. Overall, both meta-analyses must be interpreted with significant caution given methodological limitations that include the merging of estimates of studies that may not be similar. The pooling of different types of probiotics may underestimate or overestimate the true effect given the heterogeneity of the different studies. Accordingly, this highlights the need for more robust evidence regarding the effect of individual bacterial strains on the immune response and disease outcomes.
Bertelson et al. [55••] studied over 40,000 children examining the effects of probiotic milk consumption during pregnancy and childhood on atopic eczema, rhinoconjunctivitis, and asthma. Compared with babies born to mothers who did not consume probiotic milk, babies whose mothers did drink probiotic milk had reduced risk for eczema (adjusted RR 0.94, 95 % CI 0.89–0.99) at 6 months, although this link was not sustained at 18 months unless the child continued probiotic consumption (Table 2). Probiotics in pregnancy also reduced the risk for rhinoconjunctivitis (RR 0.87, 95 % CI 0.78–0.98) at 36 months of age, a risk further reduced if the infant also drank probiotic milk (Table 2) [49]. Consistent with the results of Elazab et al. [53] there was no association between probiotic milk consumption and asthma (Table 2) [55••]. It is worth highlighting that the probiotic milk products utilized in this study contained a mixture of organisms including L. acidophilus previously associated with increased risk for atopic sensitization [53].
While numerous studies associate differences in gut microbiota with weight, there is less evidence supporting the effects of early probiotics on obesity later in life [56]. In a small retrospective study (n = 30), children of normal weight at 10 years had higher numbers of fecal Bifidobacteria at age 3 months compared with children who were overweight at 10 years of age [57]. Normal weight children also were exposed to increased adiponectin in maternal colostrum, so whether differences in gut microbiota cause differences in weight or only correlate with changes in metabolism remains unclear [57]. In another study, Luoto et al. [58] followed the weight of 113 children over 10 years whose mothers consumed L. rhamnosus GG 4 weeks before delivery and then who received probiotics during the first 6 weeks of life. When adjusted for birth weight, a trend emerged where children exposed to probiotics early weighed less than those in the placebo group. The effect was more pronounced for overweight children although the subgroup was small (n = 25) [58]. A meta-analysis of 17 randomized controlled human trials, 51 farm animal studies and 14 experimental models, examined the effect of different Lactobacillus sp. administration on weight [59]. The study found that L. gasseri was associated with weight loss in obese adults while L. acidophilus was associated with weight gain [59]. No consistent effect of L. rhamnosus, a common probiotic used in multiple pediatric studies, on weight was identified [59, 60]. However, this meta-analysis must be interpreted with caution given the pooling of human and animal data, a methodological limitation given aforementioned concerns about heterogeneity. Overall, whether there is a role for early probiotic administration in obesity prevention remains to be determined and will likely depend on the specific strain of microorganism consumed.
Fecal Microbiota Transplantation
Although there has been some promising probiotic research in children, there has been an understandable gap in research related to therapeutic modulation of the microbiota at the ecological scale in the pediatric population. Fecal microbiota transplant (FMT) is a treatment strategy that restores diversity to the intestinal microbiota and has been successful in conditions where probiotics have limited efficacy such as recurrent CDI. In adults, systematic review evidence supported a clinical cure rate of approximately 90 % in recurrent CDI with particular interest in FMT via enema [61•, 62]. Despite some methodological limitations, Van Nood et al. [63••] reported robust success in the first randomized controlled study for recurrent CDI, which was stopped early for benefit. FMT via duodenal infusion yielded an 81 % (13/16 patients) clinical cure rate compared with only 31 % (4/13 patients) for vancomycin alone and 23 % (3/13 patients) for vancomycin with bowel lavage (p < 0.001 for both comparisons with the FMT group) [63••]. Similar success has been reported in a randomized controlled trial of 18 men with metabolic syndrome who underwent FMT from a lean donor or autologous FMT of their own stool [64]. These randomized studies highlight the promise of novel microbiota-directed therapies for both GI and non-GI disease in the future.
However, we must proceed with caution before applying FMT to the pediatric population. As outlined previously, there are dynamic fluctuations early in life with complex, linked microbial communities [65, 66]. Accordingly, disequilibrium early in life by microbiota-directed therapy may have downstream consequences, with the potential for adverse events. At this point, FMT must be navigated cautiously particularly in the pediatric population given the introduction of infectious disease or persistent phenotypic changes to the host [4, 61•]. Although there are various donor selection criteria reported [67, 68], there are noteworthy omissions regarding obesity and neuro-psychiatric metrics. Additionally, the literature is not consistently reporting long-term follow up for adverse events (3 weeks–8 years), which have been spontaneously reported rather than actively sought, increasing the risk for underreporting [61•]. Theoretical speculation exists regarding adverse events including peritonitis, enteritis and GI bleeding; however, these are linked to the mode of delivery and not FMT itself. More importantly, infectious, metabolic and neuro-psychiatric adverse events have not been routinely followed. Ultimately, long-term safety registries and extended follow up from emerging prospective randomized controlled trials are required before ubiquitous FMT application in the pediatric population. Despite these limitations, FMT remains promising as a potential novel therapeutic approach and warrants further exploration in this evolving field.
Conclusions
Although the field is still rapidly evolving and study outcomes are at times divergent, the composition and diversity of the gut microbiota in children is increasingly recognized as an integral component of health and disease. Evidence is also accumulating that the microbiota that colonize the GI tract in early childhood have a downstream impact on health later in life. Although exploring this intricate relationship remains in its infancy, it invites the exciting possibility that manipulating childhood gut microbiota may produce lasting health benefits. Pediatric microbiome research must continue to examine first the dynamic ‘healthy’ microbiome during childhood and changes in the microbiota that result in disease before meaningfully exploring microbiota-derived therapies. A focus on microbial–host interactions and metabolic properties of specific bacterial strains will be critical to identifying organisms most likely to have therapeutic benefit, or alternatively moving towards a systems biology solution with multiple microorganisms. The safety of children is also paramount in conducting clinical research trials so the long-term safety profile of these microbial interventions must be carefully monitored. Despite this caution, research into the pediatric microbiome remains promising and has the potential to offer novel therapies for certain pediatric diseases and, equally important, to prevent disease later in life.
Abbreviations
- Microbiota:
-
Describes vast community of microbes that colonize the human body
- Microbiome:
-
Collective genetic repertoire (genome and gene products) of microbiota
- Metagenomics:
-
Field that serves to study the relationship between genomes within intricate microbial communities
- Commensal:
-
Non-pathological relationship between two organisms, traditionally where one organism benefits and the other is not significantly helped or harmed
- Dysbiosis:
-
Imbalance or disruption of a microbial community, which may lead to pathology
- Enterotype:
-
Core group of microbes, predominantly driven by a prominent bacterial genera
- Phylotype:
-
Relationships of one microorganism to another determined by comparing 16S rRNA gene sequences. A common threshold used to define species-level phylotype is 97 % sequence identity of the 16S rRNA gene sequence
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8.
Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi:10.1038/nature11550.
Kim BS, Jeon YS, Chun J. Current status and future promise of the human microbiome. Pediatr Gastroenterol Hepatol Nutr. 2013;16:71–9.
Collins SM, Kassam Z, Bercik P. The adoptive transfer of behavioral phenotype via the intestinal microbiota: experimental evidence and clinical implications. Curr Opin Microbiol. 2013;16:240–5.
Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition in a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–8.
Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90.
Nauta AJ, Ben-Amor K, Knol J, et al. Relevance of pre- and postnatal nutrition to development and interplay between the microbiota and metabolic and immune systems. Am J Clin Nutr. 2013;98(Suppl):586–93.
Kuo CH, Kuo HF, Huang CH, et al. Early life exposure to antibiotics and the risk of childhood allergic diseases: an update from the perspective of the hygiene hypothesis. J Microbiol Immunol Infect. 2013;46(5):320–9.
Turngaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–10.
Arumugam M, Raes J, Pelletier E, et al.; MetaHIT Consortium. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80.
•• Fraher MH, O’Toole PW, Quigley EMM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol. 2012;9:312–22. This is an excellent detailed review and critique of the many techniques currently available to extract and analyze data regarding microbiota in the gut.
Arumugam M, Raes J M, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80.
Azad MB, Konya T, Maughan H, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–94.
Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012;125(5):950–60.
Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2013;148:1258–70.
Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197(3):435–8.
Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57.
de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(Suppl 1):S45–56.
Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4.
Vaahtovuo J, Munukka E, Korkeamaki M, et al. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35(8):1500–5.
Gosalbes MJ, Llop S, Valles Y, et al. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy. 2013;43:198–211.
Hu J, Nomura Y, Bashir A, et al. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE. 2013;8(11):e78257. doi:10.1371/journal.pone.0078257.
Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–5.
Breitbart B, Haynes M, Kelley S, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–73.
Vaishampayan PA, Kuehl JV, Froula JL, et al. Comparative metagenomics and population dynamics of he gut microbiota in mothers and infants. Genome Biol Evol. 2010;2:53–66.
Thum C, Cookson AL, Otter D, et al. Can nutritional modulation of maternal intestinal microbiota influence the development of the infant gastrointestinal tract? J Nutr. 2012;142(11):1921–8.
Collado MC, Cernada M, Bäuerl C, et al. Microbial ecology and host–microbiota interactions during early life. Gut Microbes. 2012;3(4):352–65.
Fallini M, Amarri S, Uusijarvi A, et al. Determinants of the human intestinal microbiota after the introduction of first complementary foods in infant samples from European centres. Microbiology. 2011;157:1385–92.
Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Supp1):4578–85.
Sartor RB, Muehlbauer M. Microbial host interaction in IBD: implications for pathogenesis and therapy. Curr Gastroenterol Rep. 2007;9(6):497–507.
Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe. World J Gastroenterol. 2011;17(5):557–66.
Vanderploeg R, Panaccione R, Ghosh S, Rioux K. Influences of intestinal bacteria in human inflammatory bowel disease. Infect Dis Clin N Am. 2010;24(4):977–93.
Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2011;1:179–84.
Sha S, Xu B, Wang X, et al. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn Microbiol Infect Dis. 2013;75:245–51.
Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Michail S, Durbin M, Turner D, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18(10):1799–808.
• Kronman MP, Zaoutis TE, Haynes K, et al. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics. 2012;130:e794–803. This is an important observational study linking antibiotic exposure and IBD highlighting the need for appropriate antibiotic stewardship.
Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3.
Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. 2011;62:361–80.
•• Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341. doi:10.1126/science.1241214. This human–mouse model study is important as it elegantly illustrates the causal effect of the microbiome on obesity.
Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–23.
Reinhardt C, Reigstad CS, Backhed F. Intestinal microbiota during infancy and its implications for obesity. JPGN. 2009;48:249–56.
Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31.
Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. 2008;87:534–8.
Xu P, Li M, Zhang J, Zhang T. Correlation of intestinal microbiota with overweight and obesity in Kazakh school children. BMC Microbiol. 2012;12:283. doi:10.1186/1471-2180-12-283.
Ismail IH, Oppedisano F, Joseph SJ, et al. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr Allergy Immunol. 2012;23:674–81.
Bisgaard H, Li N, Bonnelykke K, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128(3):646–52.
Abrahamsson TR, Jakobsson HE, Andersson AF, et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2013. doi:10.1111/cea.12253 (Epub ahead of print).
Stensballe LG, Simonsen J, Jensen SM, et al. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. J Pediatr. 2013;162:832–8.
•• Hoskin-Parr L, Teyhan A, Blocker A, Henderson AJW. Antibiotic exposure in the first two years of life and development of asthma and other allergic diseases by 7.5 yr: a dose-dependent relationship. Pediatr Allergy Immunol. 2013;24:762–71. The dose dependent relationship between antibiotic exposure and asthma and allergic disease is striking in this cohort study of 4952 children.
FAO/WHO. Guidelines for the evaluation of probiotics in food. Joint FAO/WHO Working Group Report on Drafting for the Evaluation of Probiotics in Food. 2002. www.who.int/foodsafety/publications/fs_management/probiotics2. Accessed 14 Dec 2013.
Ismail IH, Oppedisano F, Joseph SJ, et al. Prenatal administration of Lactobacillus rhamnosus has no effect on the diversity of the early infant gut microbiota. Pediatr Allergy Immunol. 2012;23:255–8.
Elazab N, Mendy A, Gasana J, et al. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. 2013;132(3):e666–76.
Pelucchi C, Chatenoud L, Turati F, et al. Probiotics supplementation during pregnancy or infancy for the prevention of atopic dermatitis: a meta-analysis. Epidemiology. 2012;23(3):402–14.
•• Bertelsen R, Brantsæter J, Magnus AL, et al. Probiotic milk consumption in pregnancy and infancy and subsequent childhood allergic diseases. J Allergy Clin Immunol. 2013. doi:10.1016/j.jaci.2013.07.032 (Epub ahead of print). This is a large cohort study of over 40,000 maternal-child pairs providing evidence for early probiotic use and protection against later allergic disease.
Arora T, Singh S, Sharma RK. Probiotics: interaction with gut microbiome and anti-obesity potential. Nutrition. 2013;29(4):591–6.
Luoto R, Kalliomäki M, Laitinen K, et al. Initial dietary and microbiological environments deviate in normal-weight compared to overweight children at 10 years of age. J Pediatr Gastroenterol Nutr. 2011;52(1):90–5.
Luoto R, Kalliomäki M, Laitinen K, Isolauri E. The impact of perinatal probiotic intervention on the development of overweight and obesity: follow-up study from birth to 10 years. Int J Obes. 2010;34(10):531–7.
Million M, Angelakis E, Paul M, et al. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb Pathog. 2012;53(2):100–8.
Million M, Lagier JC, Yahav D, Paul M. Gut bacterial microbiota and obesity. Clin Microbiol Infect. 2013;19(4):305–13.
• Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol. 2013;108:500–8. This is an important meta-analysis that robustly summarizes the impact of fecal microbiota transplantation in Clostridium difficile infection and explores areas of concern, including safety in this field.
Kassam Z, Hundal R, Marshall JK, Lee CH. Fecal transplant via retention enema for refractory or recurrent Clostridium difficile infection. Arch Intern Med. 2010;172:191–3.
•• Van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile infection. N Engl J Med. 2013;368:407–15. This is the first randomized controlled trial for fecal microbiota transplant for Clostridium difficile, and it was stopped early for benefit highlighting the potential of this emerging therapy.
Vreize A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donor increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–6.
Palmer C, Bik EM, Digiulio DB, et al. Development of human infant intestinal microbiota. PLoS Biol. 2007;5(e177):1556–73.
Tiihonen K, Ouwehand AC, Rautonen N. Human intestinal microbiota and healthy ageing. Ageing Res Rev. 2010;9(2):107–16.
Allen-Vercoe E, Reid G, Viner N, et al. Canadian Working Group report on fecal microbiota therapy: microbial ecosystem therapeutics. Can J Gastroenterol. 2012;26(7):457–62.
Bakken JS, Borody T, Brandt LJ, et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2011;9(12):1044–9.
Disclosures
Zain Kassam and Thomas S. Murray declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kassam, Z., Murray, T.S. Navigating the Pediatric Microbiome: Emerging Evidence and Clinical Implications. Curr Pediatr Rep 2, 93–101 (2014). https://doi.org/10.1007/s40124-014-0040-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40124-014-0040-1