Avoid common mistakes on your manuscript.
Due to the rapid spread and increasing number of coronavirus disease 19 (COVID-19) cases caused by Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), simple, rapid, sensitive, and specific technique is necessary for routine detection of virus in laboratories and early diagnosis of COVID-19 is vital to prevent and control of this pandemic [1]. The detection of viral nucleic acid is the standard way for the noninvasive diagnosis of COVID-19. There are several molecular techniques for the detection of SARS-CoV-2 based on viral nucleic acid in respiratory samples [2]. Real-time RT-PCR on nasopharyngeal and oropharyngeal swabs is considered as the “gold standard” for confirming the diagnosis in clinical cases of COVID-19, which entails one or several primer–probe sets for targeting SARS-CoV-2 sequences [3]. The primer–probe sets target the different regions of SARS-CoV-2 including; orf1 (a, b), envelope (E), nucleocapsid (N), spike (S), and RNA-dependent RNA polymerase (RdRp) sequences as the target of the RT-PCR, showing that each of the genes has different sensitivity and specificity, and they can be used for detection and confirming of SARS-CoV-2 worldwide [4]. In early response and detection of the SARS-COV-2 (COVID-19) outbreak, the cause of the recent quickly spreading respiratory tract infection, the World Health Organization (WHO) recommended an RT-PCR first-line screening test based on an E-gene assay, followed by a confirmatory assay using the RdRp gene, since RT-PCR assays targeting the RdRp assay had the highest analytical sensitivity [5]. Furthermore, it was demonstrated that the use of two primer–probe sets targeting the nucleocapsid gene (N1 and N2) as suggested by the Centers of Disease Control and Prevention (CDC), will help to facilitate sensitive and specific detection of SARS-CoV-2 in the future [6]. Most studies used at least two primer–probe sets such as N and RdRp or N1 and N2 for the detection of COVID-19 infections [7,8,9]. Regarding the fact that primer-probes sets are considered as a critical factor for accurate detection of SARS-CoV-2 in clinical samples, choosing the best target gene in commercial RT-PCR methods appears to be necessary. In this brief report, we analyzed amplification curves of the two target genes and highlighted the pitfalls of these targets. Additionally, we compared the sensitivity and specificity of these genes for the detection of COVID-19 infections.
In this study, 114 respiratory specimens (nasopharyngeal and oropharyngeal swabs) collected from symptomatic patients admitted to various medical centers were immediately placed into sterile tubes containing 3 mL of viral transport media (VTM). The medium constitutes Hank’s balanced salt solution at pH 7.4 containing BSA (1%), amphotericin (15 μg/mL), penicillin G (100 units/mL), and streptomycin (50 μg/mL). Samples (3 ml) were kept at 4–8 °C for short-term storage and at − 70 °C for the long term.
Viral RNA was extracted from 300 µL of nasopharyngeal and oropharyngeal swabs of suspected COVID-19 patients using an RNA extraction kit (Gene All Biotechnology, Seoul, South Korea), and eluted in 50 µL of nuclease-free water. RNA extracts were stored at − 70 °C for further analyses. For detection of SARS-CoV-2, RT-qPCR primer–probe sets designed on RdRp and N genes that were introduced by World Health Organization (WHO) were used. For all samples, RT-qPCR was performed for N and RdRp genes using the one-step RT-qPCR kit (Sansure Biotech, Changsha/Hunan, China), according to the manufacturer’s instructions. Additionally, the RNase P gene was used as an internal control for monitoring the validity of sample collection, and RT-qPCR process to avoid false-negative results. Thermal cycle conditions were as follows: 30 min at 50 °C for reverse transcription, 1 min at 95 °C for PCR initiation activation, and 45 cycles of 95 °C at 15 s and 30 s at 60 °C. The primer–probe set sequences (Table 1) and their position on the SARS-COV-2 genome for N and RdRp segments are shown in Fig. 1.
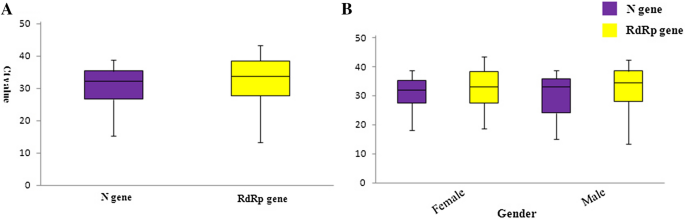
Patients were only evaluated by gender, not further characteristics, and by their Ct values no correlation with age is given, and Ct values for N- and RdRp gene amplification number, as shown in the supplementary Table. Of the 114 patients, 64 (56.14%) and 50 (43.86%) were female and male, respectively. According to the Chi-square test and P value (P = 0.8), no significant difference was observed between gender and test results. The results of the amplification plot for each clinical sample showed (the amplification plots are shown as supplementary figure) that there was a significant positive signal of Ct values finishing the SARS-CoV-2N- and RdRp-gene RT-PCR amplification. To estimate amplification efficiency by ROC curve (Receiver Operating Characteristic), Ct values were analyzed at the end of the RT-PCR program. Results from triplicate analyses were and the mean was plotted as Ct value (Fig. 2). The results of the area under the ROC curve showed that the N Ct value (value = 0.980), was more specific in the detection of SARS-CoV-2 compared to the RdRp Ct value (value = 0.647) (Fig. 3).
Comparative analysis of Ct values for N- and RdRp primer–probe sets from clinical samples. A N- and RdRp-genes Ct value analysis of SARS-CoV-2 detection in the clinical samples showed that the N-gene according to the Ct value appearance is one of the favorable targets for amplification. B The sex group analysis of SARS-CoV-2 positive cases revealed that no significant difference was observed between gender and Ct value distribution
Real-time RT-PCR as “gold standard” and nucleic acid detection-based technique is considered in the global response to the COVID-19 pandemic for detection of viral RNAs in clinical samples such as sputum, nasal swab, and throat swab [10]. Along with the rapidly increasing number of COVID-19 cases, sensitive, and specific detection of virus in infected patients has been key in controlling the outbreak. It has been well known that results from real-time RT-PCR can be affected by several parameters including the (1) primer/probe set, (2) sample quantity or viral load, (3) quality and type of sample such as the anatomic site testing (mouth, nasopharynx or BAL (broncho-alveolar lavage)), time of sampling [11]. Primer and probe set is the most common cause of false-negative results in real-time PCR-based protocols. In this regard, multiple amplification sets are the most widely used for the detection of SARS-CoV-2, to avoid false-negative results [12]. Several types of primer and probe sets have been optimized and designed rapidly, but with different sensitivity and specificity. An important issue with the primer–probe set is the lack of suitable analytical sensitivity and specificity, and PCR amplification efficiency. The N gene (nucleocapsid) is one of the best and most accurate targets for detecting, tracking, and first-line screening of the SARS-CoV-2, based on the WHO and CDC guidelines, commercial kits, and scientific evaluations [13].
The present report demonstrates that the N primer and probe set has a higher specificity compared to the RdRp primer and probe set, and also the superior method for identifying new cases of SARS-CoV-2 in clinical samples, based on the real-time PCR Ct value. Optimization of RT-qPCR methods that are using one primer–probe set can reduce diagnostic costs. Additionally, in the multiplex-qPCR method for simultaneous detection and differentiation of SARS-CoV-2 and other respiratory viruses, there are limitations associated with the number of used primer–probe sets. Hence, we have to use one primer–probe set for SARS-CoV-2 detection. Taken together, selecting primer–probe sets for two viral genes that yield acceptable real-time PCR Ct values is increasingly vital in simplex and multiplex RT-qPCR protocols. The limitation of the present study was no investigation on the sensitivity and PCR amplification efficiency of the RdRp and N primer–probe sets, by testing for example tenfold dilutions of the synthetic SARS-CoV-2 diagnostic RNA. Furthermore, analytical factors including, sensitivity, specificity, PCR amplification efficiency, and Ct value affected the choice of primer–probe sets, the latter of which was analyzed in the present study [13].
References
Harapan H, Itoh N, Yufika A, Winardi W, Keam S, Te H, et al. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020;13:667–73.
Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J Clin Med. 2020;9:1225.
Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, et al. Coronavirus disease 2019–COVID-19. Clin Microbiol Rev. 2020;33:e00028-e00120.
Tang Y-W, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58:e00512-e520.
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045.
Centers for Disease Control and Prevention. The CDC 2019 novel coronavirus (2019 nCoV) real time Rt-PCR diagnostics. https://www.fda.gov/media/134922/download.pdf (2019).
Mollaei HR, Afshar AA, Kalantar-Neyestanaki D, Fazlalipour M, Aflatoonian B. Comparison five primer sets from different genome region of COVID-19 for detection of virus infection by conventional RT-PCR. Iran J Microbiol. 2020;12:185.
Barra GB, Santa Rita TH, Mesquita PG, Jácomo RH, Nery LFA. Analytical sensitivity and specificity of two RT-qPCR protocols for SARS-CoV-2 detection performed in an automated workflow. Genes. 2020;11:1183.
Etievant S, Bal A, Escuret V, Brengel-Pesce K, Bouscambert M, Cheynet V, et al. Performance assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J Clin Med. 2020;9:1871.
Oliveira BA, Oliveira LCD, Sabino EC, Okay TS. SARS-CoV-2 and the COVID-19 disease: a mini review on diagnostic methods. Rev Inst Med Trop São Paulo. 2020;62:e44.
Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J Adv Res. 2020;26:149–59.
Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20:453–4.
Vogels CB, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer–probe sets. Nat Microbiol. 2020;5:1299–305.
Acknowledgements
We are warmly grateful to all those who were involved in this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicting interests.
Ethical consideration
This project is based on adhering to the declaration of Helsinki of 2013 and was approved by the Ethics Committee of Golestan University of Medical Sciences (Ethics code: IR.GOUMS.REC.1399.432).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abbasi, H., Tabaraei, A., Hosseini, S.M. et al. Real-time PCR Ct value in SARS-CoV-2 detection: RdRp or N gene?. Infection 50, 537–540 (2022). https://doi.org/10.1007/s15010-021-01674-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-021-01674-x