Abstract
BACKROUND:
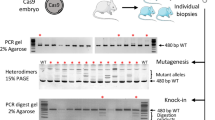
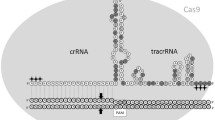
CRISPR/Cpf1 is a class II, type V RNA-guided endonuclease that is distinct from the type II CRISPR/Cas9 nuclease, widely used for genome editing. Cpf1 is a smaller and simpler endonuclease than Cas9, overcoming some limitations of the CRISPR/Cas9 system. The applications of CRISPR to rodent embryos for the production of knock-out (KO) mice have been achieved mainly by microinjection, which requires heavily-equipped instruments with skillful hands. Here, we evaluated the genome editing efficiency between Cpf1/mRNA and Cpf1/ribonuclear protein (RNP) in mouse embryos, and established an easy, fast, and technically less demanding method to produce KO mice using electroporation of the Cfp1/RNP system.
METHODS:
The efficiency of electroporation-based delivery of AsCpf1/mRNA and AsCpf1/RNP to target exon 3 of leukemia inhibitory factor (Lif) into mouse zygotes was evaluated. Embryos that developed to the two-cell stage after zygote electroporation were transferred into the oviducts of surrogate mothers to produce AsCpf1-mediated LIF KO mice. The genome editing efficiency of blastocysts and pups was tested using the T7E1 assay and/or DNA sequencing. Congenital abnormalities and reproductive phenotypes in LIF KO mice produced by electroporation with AsCpf1/RNP were examined.
RESULTS:
Survival and two-cell development of electroporated zygotes were comparable between the AsCpf1/mRNA and AsCpf1/RNP groups, whereas genome editing efficiency was relatively higher in the AsCpf1/RNP group (13.3% vs 18.1% at blastocyst and 33.3% vs 45.5% at offspring), respectively. Two mouse lines with a frameshift mutation in exon 3 of the Lif gene were established from the AsCpf1/RNP group. All congenital abnormalities of LIF KO mice produced by AsCpf1/RNP electroporation were observed. AsCpf1-mediated LIF KO mice showed postnatal growth retardation and implantation failure, both of which are major phenotypes of LIF KO mice generated by conventional gene targeting.
CONCLUSION:
Electroporation of AsCpf1/RNP at the zygote stage is an efficient genome editing method to produce KO mice.




Similar content being viewed by others
References
Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–12.
Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, et al. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31:23–4.
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8.
Wefers B, Ortiz O, Wurst W, Kühn R. Generation of targeted mouse mutants by embryo microinjection of TALENs. Methods. 2014;69:94–101.
Chen YG, Forsberg MH, Khaja S, Ciecko AE, Hessner MJ, Geurts AM. Gene targeting in NOD mouse embryos using zinc-finger nucleases. Diabetes. 2014;63:68–74.
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–9.
Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6.
Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–9.
Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53.
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell. 2015;163:759–71.
Kim Y, Cheong SA, Lee JG, Lee SW, Lee MS, Baek IJ, et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016;34:808–10.
Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863–8.
Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, et al. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–4.
Watkins-Chow DE, Varshney GK, Garrett LJ, Chen Z, Jimenez EA, Rivas C, et al. Highly efficient Cpf1-mediated gene targeting in mice following high concentration pronuclear injection. G3 (Bethesda). 2017;7:719–22.
Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q, et al. Repurposing CRISPR–Cas12b for mammalian genome engineering. Cell Discov. 2018;4:63.
Lee JG, Ha CH, Yoon B, Cheong SA, Kim G, Lee DJ, et al. Knockout rat models mimicking human atherosclerosis created by Cpf1-mediated gene targeting. Sci Rep. 2019;9:2628.
Wu H, Liu Q, Shi H, Xie J, Zhang Q, Ouyang Z, et al. Engineering CRISPR/Cpf1 with tRNA promotes genome editing capability in mammalian systems. Cell Mol Life Sci. 2018;75:3593–607.
Moreno-Mateos MA, Fernandez JP, Rouet R, Vejnar CE, Lane MA, Mis E, et al. CRISPR–Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun. 2017;8:2024.
Chen S, Lee B, Lee AY, Modzelewski AJ, He L. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J Biol Chem. 2016;291:14457–67.
Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol. 2016;34:807–8.
Wang W, Kutny PM, Byers SL, Longstaff CJ, DaCosta MJ, Pang C, et al. Delivery of Cas9 protein into mouse zygotes through a series of electroporation dramatically increases the efficiency of model creation. J Genet Genomics. 2016;43:319–27.
Lin Y, Cradick TJ, Bao G. Designing and testing the activities of TAL effector nucleases. In: Storici F, editor. Gene Correction. Methods in Molecular Biology (Methods and Protocols), Vol 1114. Humana Press, Totowa, NJ; 2014. p. 203–19.
Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–6.
Kissling L, Monfort A, Swarts DC, Wutz A, Jinek M. Preparation and electroporation of Cas12a/Cpf1-guide RNA complexes for introducing large gene deletions in mouse embryonic stem cells. Methods Enzymol. 2019;616:241–63.
Bin Moon S, Lee JM, Kang JG, Lee NE, Ha DI, Kim DY, et al. Highly efficient genome editing by CRISPR–Cpf1 using CRISPR RNA with a uridinylate-rich 3′-overhang. Nat Commun. 2018;9:3651.
Hilton DJ, Gough NM. Leukemia inhibitory factor: a biological perspective. J Cell Biochem. 1991;46:21–6.
Schäfer-Somi S. Cytokines during early pregnancy of mammals: a review. Anim Reprod Sci. 2003;75:73–94.
Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20.
Bhatt H, Brunet LJ, Stewart CL. Uterine expression of leukemia inhibitory factor coincides with the onset of blastocyst implantation. Proc Natl Acad Sci U S A. 1991;88:11408–12.
Rosario GX, Stewart CL. The multifaceted actions of leukaemia inhibitory factor in mediating uterine receptivity and embryo implantation. Am J Reprod Immunol. 2016;75:246–55.
Song H, Lim H, Das SK, Paria BC, Dey SK. Dysregulation of EGF family of growth factors and COX-2 in the uterus during the preattachment and attachment reactions of the blastocyst with the luminal epithelium correlates with implantation failure in LIF-deficient mice. Mol Endocrinol. 2000;14:1147–61.
Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Köntgen F, et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–9.
Escary JL, Perreau J, Duménil D, Ezine S, Brûlet P. Leukaemia inhibitory factor is necessary for maintenance of haematopoietic stem cells and thymocyte stimulation. Nature. 1993;363:361–4.
Charnock-Jones DS, Sharkey AM, Fenwick P, Smith SK. Leukaemia inhibitory factor mRNA concentration peaks in human endometrium at the time of implantation and the blastocyst contains mRNA for the receptor at this time. J Reprod Fertil. 1994;101:421–6.
Chen JR, Cheng JG, Shatzer T, Sewell L, Hernandez L, Stewart CL. Leukemia inhibitory factor can substitute for nidatory estrogen and is essential to inducing a receptive uterus for implantation but is not essential for subsequent embryogenesis. Endocrinology. 2000;141:4365–72.
Kimber SJ. Leukaemia inhibitory factor in implantation and uterine biology. Reproduction. 2005;130:131–45.
Stewart CL. Leukaemia inhibitory factor and the regulation of pre-implantation development of the mammalian embryo. Mol Reprod Dev. 1994;39:233–8.
Acknowledgements
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2019R1A6A1A03032888) and from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant No. HI17C1133).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical statement
All mice used in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of CHA University (IACUC approval No. IACUC170174).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, Y.S., Kim, G.R., Park, M. et al. Electroporation of AsCpf1/RNP at the Zygote Stage is an Efficient Genome Editing Method to Generate Knock-Out Mice Deficient in Leukemia Inhibitory Factor. Tissue Eng Regen Med 17, 45–53 (2020). https://doi.org/10.1007/s13770-019-00225-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13770-019-00225-8