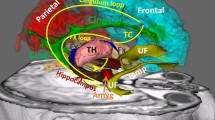
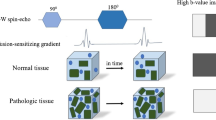
Abstract
Diffusion magnetic resonance imaging (dMRI) is a cutting-edge imaging method that provides a macro-scale in vivo map of the white matter pathways in the brain. The measurement of brain microstructure and the enhancement of tractography rely heavily on dMRI tissue segmentation. Anatomical MRI technique (e.g., T1- and T2-weighted imaging) is the most widely used method for segmentation in dMRI. In comparison to anatomical MRI, dMRI suffers from higher image distortions, lower image quality, and making inter-modality registration more difficult. The dMRI tractography study of brain connectivity has become a major part of the neuroimaging landscape in recent years. In this research, we provide a high-level overview of the methods used to segment several brain tissues types, including grey and white matter and cerebrospinal fluid, to enable quantitative studies of structural connectivity in the brain in health and illness. In the first part of our review, we discuss the three main phases in the quantitative analysis of tractography, which are correction, segmentation, and quantification. Methodological possibilities are described for each phase, along with their popularity and potential benefits and drawbacks. After that, we will look at research that used quantitative tractography approaches to examine the white and grey matter of the brain, with an emphasis on neurodevelopment, ageing, neurological illnesses, mental disorders, and neurosurgery as possible applications. Even though there have been substantial advancements in methodological technology and the spectrum of applications, there is still no consensus regarding the "optimal" approach in the quantitative analysis of tractography. As a result, researchers should tread carefully when interpreting the findings of quantitative analysis of tractography.





Similar content being viewed by others
Data Availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Change history
23 June 2023
A Correction to this paper has been published: https://doi.org/10.1007/s13760-023-02301-2
References
Hasan A, Meziane F, Aspin R, Jalab H (2016) Segmentation of brain tumors in MRI images using three-dimensional active contour without edge. Symmetry 8(11):132
Gordillo N, Montseny E, Sobrevilla P (2013) State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging 31(8):1426–1438
Bahadure N, Kumar Ray A, Pal TH (2017) Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. Int J Biomed Imaging 2017:1–12
Wong KP (2005) Medical image segmentation: methods and applications in functional imaging. In: Suri JS, Wilson DL, Laxminarayan S (eds) Handbook of biomedical image analysis. Topics in biomedical engineering international book series. Springer, Boston. https://doi.org/10.1007/0-306-48606-7_3
Jeurissen B, Tournier J-D, Sijbers J (2015) Tissue-type segmentation using non-negative matrix factorization of multi-shell diffusion-weighted MRI images. In: ISMRM 23th Annual Meeting, Toronto, Ontario, Canada, volume 23, p. 349
Kumazawa S, Yoshiura T, Honda H, Toyofuku F, Higashida Y (2010) Partial volume estimation and segmentation of brain tissue based on diffusion tensor MRI. Med Phys 37(4):1482–1490
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
CBICA Homepage, https://ipp.cbica.upenn.edu/. Last accessed 10 May 2022
Mah Y-H, Jager R, Kennard C, Husain M, Nachev P (2014) A new method for auto- mated high-dimensional lesion segmentation evaluated in vascular injury and applied to the human occipital lobe. Cortex 56:51–63
Kumar R, Rani S, Sarkar A, Talukdar FA (2017) GPU-based level set method for MRI brain tumor segmentation using modified probabilistic clustering. IGI Global, pp 1053–1078
Ilunga-Mbuyamba E, Avina-Cervantes JG, Garcia-Perez A, de Jesus-Romero-Troncoso R, Aguirre-Ramos H, Cruz-Aceves I (2017) Localized active contour model with background intensity compensation applied on automatic MR brain tumor segmentation. Neurocomputing 220:84–97
Soltaninejad M, Zhang L, Lambrou T, Yang G, Allinson N, Ye X (2017) MRI brain tumor segmentation and patient survival prediction using random forests and fully convolutional networks. In: Int. MICCAI Brain lesion Workshop. Springer. pp 204–15
Parveen SA (2015) Detection of brain tumor in MRI images, using combination of fuzzy C-means and SVM. In: 2nd Int. Conf. Signal Processing and Integrated Networks (SPIN), pp 98–102
Soltaninejad M, Zhang L, Lambrou T, Yang G, Allinson N, Ye X (2017) MRI brain tumor segmentation using random forests and fully convolutional networks. In: International MICCAI Brain lesion Workshop, pp 279–83
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats’ challenge. arXiv preprint https://arXiv.org/181102629
Steven AJ, Zhuo J, Melhem ER (2014) Diffusion kurtosis imaging: an emerging technique for evaluating the microstructural environment of the brain. Am J Roentgenol 202(1):26–33
Sun P, Wu Y, Chen G, Wu J, Shen D, Yap P-T (2019) Tissue segmentation using sparse non-negative matrix factorization of spherical mean diffusion MRI data. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp 69–76
Tabesh A, Jensen JH, Ardekani BA, Helpern JA (2011) Estimation of tensors and tensor-derived measures in diffusional kurtosis imaging. Magn R83eson Med 65(3):823–836
Tong Q, He H, Gong T, Li C, Liang P, Qian T, Sun Y, Ding Q, Li K, Zhong J (2020) Multicenter dataset of multi-shell diffusion MRI in healthy traveling adults with identical settings. Sci Data 7(1):1–7
Tournier J-D, Calamante F, Connelly A (2007) Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35(4):1459–1472
Veraart J, Fieremans E, Jelescu IO, Knoll F, Novikov DS (2016) Gibbs ringing in diffusion MRI. Magn Reson Med 76(1):301–314
Wasserthal J, Neher P, Maier-Hein KH (2018) Tractseg-fast and accurate white matter tract segmentation. Neuroimage 183:239–253
Wen Y, He L, von Deneen KM, Lu Y (2013) Brain tissue classification based on DTI using an improved fuzzy C-means algorithm with spatial constraints. Magn Reson Imaging 31(9):1623–1630
Wu M, Chang L-C, Walker L, Lemaitre H, Barnett AS, Marenco S, Pierpaoli C (2008) Comparison of EPI distortion correction methods in diffusion tensor mri using a novel framework. Medical image computing and computer-assisted intervention. Springer, pp 321–329
Yap P-T, Zhang Y, Shen D (2015) Brain tissue segmentation based on diffusion MRI using L0 sparse-group representation classification. Medical image computing and computer-assisted intervention. Springer, pp 132–139
Zhang F, Cho KIK, Tang Y, Zhang T, Kelly S, Di Biase M, Xu L, Li H, Matcheri K, Whitfield-Gabrieli S et al (2020) MK-curve improves sensitivity to identify white matter alterations in clinical high risk for psychosis. Neuroimage 226:117564
Zhang F, Ning L, O’Donnell LJ, Pasternak O (2019) MK-curve—characterizing the relation between mean kurtosis and alterations in the diffusion MRI signal. Neuroimage 196:68–80
Zhang F, Noh T, Juvekar P, Frisken SF, Rigolo L, Norton I, Kapur T, Pujol S, Wells W, Yarmarkovich A, Kindlmann G, Wassermann D, San-Jose-Estepar R, Rathi Y, Kikinis R, Johnson HJ, Westin C-F, Pieper S, Golby AJ, O’Donnell LJ (2020) SlicerDMRI: diffusion MRI and tractography research software for brain cancer surgery planning and visualization. JCO Clin Cancer Inform. 4:299–309
Sakkalis V (2011) Review of advanced techniques for the estimation of brain connectivity measured with EEG/MEG. Comput Biol Med 41:1110–1117
Gatys LA, Ecker AS, Bethge M (2016) Image style transfer using convolutional neural networks. In: IEEE Conference on Computer Vision and Pattern Recognition, pp 2414 – 2423
Sarubbo S, Tate M, De Benedictis A, Merler S, Moritz-Gasser S, Herbet G, Duau H (2020) Mapping critical cortical hubs and white matter pathways by direct electrical stimulation: an original functional atlas of the human brain. Neuroimage 205:116237
Sarwar T, Seguin C, Ramamohanarao K, Zalesky A (2020) Towards deep learning for connectome mapping: a block decomposition framework. Neuroimage 212:116654
Sbardella E, Tona F, Petsas N, Pantano P (2013) Dti measurements in multiple sclerosis: evaluation of brain damage and clinical implications. Mult Scler Int 2013(2013):671730
Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, Eickho SB, Yeo BT (2018) Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex 28:3095–3114
Devi CN, Chandrasekharan A, Sundararaman VK, Alex ZC (2015) Neonatal brain MRI segmentation: a review. Comput Biol Med 64:163–178
Schurr R, Filo S, Mezer AA (2019) Tractography delineation of the vertical occipital fasciculus using quantitative t1 mapping. Neuroimage 202:116121
Seguin C, van den Heuvel MP, Zalesky A (2018) Navigation of brain networks. Proc Natl Acad Sci U S A 115:6297–6302
Seguin C, Razi A, Zalesky A (2019) Inferring neural signalling directionality from undirected structural connectomes. Nat Commun 10:4289
Seguin C, Tian Y, Zalesky A (2020) Network communication models improve the behavioral and functional predictive utility of the human structural connectome. Netw Neurosci 4:980–1006
Sepasian N, ten Thije Boonkkamp J, Ter Haar Romeny B, Vilanova Bartroli A (2012) Multivalued geodesic ray-tracing for computing brain connections using diffusion tensor imaging. SIAM J Imag Sci 5:483–504
Shahab S, Stefanik L, Foussias G, Lai MC, Anderson KK, Voineskos AN (2018) Sex and diffusion tensor imaging of white matter in schizophrenia: a systematic review plus meta-analysis of the corpus callosum. Schizophr Bull 44:203–221
Pecheva D, Yushkevich P, Batalle D, Hughes E, Aljabar P, Wurie J, Hajnal JV, Edwards AD, Alexander DC, Counsell SJ et al (2017) A tract-specific approach to assessing white matter in preterm infants. Neuroimage 157:675–694
Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AM, Sestan N, Wildman DE et al (2012) Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci 109:16480–16485
Gottlieb D, Shu CW, Solomonoff A, Vandeven H (1992) On the Gibbs phenomenon I: recovering exponential accuracy from the Fourier partial sum of a nonperiodic analytic function. J Comp Appl Math 43:81–98
Misic B, Betzel RF, Nematzadeh A, Goni J, Gria A, Hagmann P, Flammini A, Ahn YY, Sporns O (2015) Cooperative and competitive spreading dynamics on the human connectome. Neuron 86:1518–1529
Mito R, Raelt D, Dhollander T, Vaughan DN, Tournier JD, Salvado O, Brodtmann A, Rowe CC, Villemagne VL, Connelly A (2018) Fibre-specific white matter reductions in Alzheimer’s disease and mild cognitive impairment. Brain 141:888–902
Cui LB, Wei Y, Xi YB, Gria A, De Lange SC, Kahn RS, Yin H, Van den Heuvel MP (2019) Connectome-based patterns of first episode medication-naive patients with schizophrenia. Schizophr Bull 45:1291–1299
Cui Z, Zhong S, Xu P, Gong G, He Y (2013) Panda: a pipeline toolbox for analyzing brain diffusion images. Front Hum Neurosci 7:42
Daducci A, Dal-Palu A, Descoteaux M, Thiran JP (2016) Microstructure informed tractography: pitfalls and open challenges. Front Neurosci 10:247
Daducci A, Dal Palu A, Lemkaddem A, Thiran JP (2013) A convex optimization framework for global tractography. In: 2013 IEEE 10th International Symposium on Biomedical Imaging, IEEE. pp 524–527
Daducci A, Dal Palu A, Lemkaddem A, Thiran JP (2014) COMMIT: convex optimization modeling for microstructure informed tractography. IEEE Trans Med Imaging 34:246–257
Damatac CG, Chauvin RJ, Zwiers MP, van Rooij D, Akkermans SE, Naaijen J, Hoekstra PJ, Hartman CA, Oosterlaan J, Franke B et al (2020) White matter microstructure in attention deficit/hyperactivity disorder: a systematic tractography study in 654 individuals. Biol Psychiatry Cogn Neurosci Neuroimaging 7:979–988
Zhanga F, Daduccib A, Yong H, Schiavib S, Seguing C, Smithi R, Yeh C-H, Zhao T, O’Donnell LJ (2021) Quantitative mapping of the brain’s structural connectivity using diffusion MRI tractography: a review. arXiv, pp 1–29
Davis SW, Dennis NA, Buchler NG, White LE, Madden DJ, Cabeza R (2009) Assessing the effects of age on long white matter tracts using diffusion tensor tractography. Neuroimage 46:530–541
De Witte NA, Mueller SC (2017) White matter integrity in brain networks relevant to anxiety and depression: evidence from the human connectome project dataset. Brain Imaging Behav 11:1604–1615
Zhang W, Olivi A, Hertig SJ, Van Zijl P, Mori S (2008) Automated fiber tracking of human brain white matter using diffusion tensor imaging. Neuroimage 42:771–777
Zhang Y, Zhang J, Oishi K, Faria AV, Jiang H, Li X, Akhter K, Rosa-Neto P, Pike GB, Evans A et al (2010) Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage 52:1289–1301
Zhang Z, Descoteaux M, Zhang J, Girard G, Chamberland M, Dunson D, Srivastava A, Zhu H (2018) Mapping population-based structural connectomes. Neuroimage 172:130–145
Zhao T, Cao M, Niu H, Zuo XN, Evans A, He Y, Dong Q, Shu N (2015) Age-related changes in the topological organization of the white matter structural connectome across the human lifespan. Hum Brain Mapp 36:3777–3792
Zhao T, Mishra V, Jeon T, Ouyang M, Peng Q, Chalak L, Wisnowski JL, Heyne R, Rollins N, Shu N et al (2019) Structural network maturation of the preterm human brain. Neuroimage 185:699–710
Zhao T, Sheng C, Bi Q, Niu W, Shu N, Han Y (2017) Age-related differences in the topological efficiency of the brain structural connectome in amnestic mild cognitive impairment. Neurobiol Aging 59:144–155
Zhao T, Xu Y, He Y (2019) Graph theoretical modeling of baby brain networks. Neuroimage 185:711–727
Zhao X, Tian L, Yan J, Yue W, Yan H, Zhang D (2017) Abnormal rich-club organization associated with compromised cognitive function in patients with schizophrenia and their unaffected parents. Neurosci Bull 33:445–454
Ziyan U, Sabuncu MR, Grimson WEL, Westin CF (2009) Consistency clustering: a robust algorithm for group-wise registration, segmentation and automatic atlas construction in diffusion MRI. Int J Comput Vision 85:279–290
Zollei L, Jaimes C, Saliba E, Grant PE, Yendiki A (2019) Tracts constrained by underlying infant anatomy (traculina): an automated probabilistic tractography tool with anatomical priors for use in the newborn brain. Neuroimage 199:1–17
Yeh CH, Smith RE, Dhollander T, Connelly A (2017) Mesh-based anatomically-constrained tractography for effective tracking termination and structural connectome construction. In: Proceedings of the ISMRM, no. 0058
Yeh CH, Smith RE, Liang X, Calamante F, Connelly A (2016) Correction for diffusion MRI fibre tracking biases: the consequences for structural connectomic metrics. Neuroimage 142:150–162
Yeh FC, Badre D, Verstynen T (2016) Connectometry: a statistical approach harnessing the analytical potential of the local connectome. Neuroimage 125:162–171
Yeh FC, Panesar S, Fernandes D, Meola A, Yoshino M, Fernandez-Miranda JC, Vettel JM, Verstynen T (2018) Population-averaged atlas of the macroscale human structural connectome and its network topology. Neuroimage 178:57–68
Urban G, Bendszus M, Hamprecht F, Kleesiek J (2014) Multimodal brain tumor segmentation using deep convolutional neural networks. In: MICCAI BraTS (Brain Tumour Segmentation) Challenge. Proceedings, Winning Contribution, pp 31–35
Vijayakumar C, Gharpure DC (2011) Development of image-processing software for automatic segmentation of brain tumors in MRI images. J Med Phys/Assoc Med Phys India 36(3):147
Wang SH, Phillips P, Sui Y, Liu B, Yang M, Cheng H (2018) Classification of Alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J Med Syst 42(5):85
Rondina JM, Ferreira LK, de Souza Duran FL, Kubo R, Ono CR, Leite CC et al (2018) Selecting the most relevant brain regions to discriminate Alzheimer’s disease patients from healthy controls using multiple kernel learning: a comparison across functional and structural imaging modalities and atlases. Neuroimage Clin 17:628–641
Saha P, Udupa J (2001) Optimum image thresholding via class uncertainty and region homogeneity. IEEE Trans Pattern Anal Mach Intell 12(7):689–706
Salman Y (2009) Modified technique for volumetric brain tumour measurements. J Biomed Sci Eng 2:16–19
Salman Y, Badawi A, Assal M, Alian S (2005) New automatic technique for tracking brain tumor response. In: International conference on biological and medical physics, pp 1–4
Sanchez A, Mammone N, Morabito FC, Marino S, Adeli H (2019) A novel methodology for automated differential diagnosis of mild cognitive impairment and the Alzheimer’s disease using EEG signals. J Neurosci Methods 322:88–95
Oxtoby NP, Garbarino S, Firth NC et al (2017) Data-driven sequence of changes to anatomical brain connectivity in sporadic Alzheimer’s disease. Front Neurol 8:580
Betzel RF, Bassett DS (2018) Specificity and robustness of long-distance connections in weighted, interareal connectomes. Proc Natl Acad Sci U S A 115:E4880–E4889
Raffelt D, Sadeghian F, Connor H. Connelly A (2015) Decreased apparent fiber density in the optic pathways correlates with glaucoma disease severity. In: Proc ISMRM, p 2213
Vaughan DN, Raffelt D, Curwood E et al (2017) Tract-specific atrophy in focal epilepsy: disease, genetics, or seizures? Ann Neurol 81:240–250
Mito R, Raffelt D, Dhollander T et al (2018) Fibre-specific white matter reductions in Alzheimer’s disease and mild cognitive impairment. Brain 141:888–902
Liang X, Yeh C-H, Connelly A, Calamante F (2019) Robust identification of rich-club organization in weighted and dense structural connectomes. Brain Topogr 32:1–16
Xing X-X, Zuo X-N (2018) The anatomy of reliability: a must read for future human brain mapping. Sci Bull 63:1606–1607
Zuo X-N, Xu T, Milham MP (2019) Harnessing reliability for neuroscience research. Nat Hum Behav 3:768–771
Tzourio-Mazoyer N, Landeau B, Papathanassiou D et al (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289
Glasser MF, Coalson TS, Robinson EC et al (2016) A multi-modal parcellation of human cerebral cortex. Nature 536:171–178
Gelb A, Tadmor E (1999) Detection of edges in spectral data. Appl Comput Harmon Anal 7:101–135
Gelb A, Tadmor E (2000) Detection of edges in spectral data II: nonlinear enhancement. SIAM J Numer Anal 38:1389–1408
Archibald R, Gelb A (2002) A method to reduce the Gibbs ringing artifact in MRI scans while keeping tissue boundary integrity. IEEE Trans Med Imaging 21:305–319
Archibald R, Chen K, Gelb A, Renautc R (2003) Improving tissue segmentation of human brain MRI through preprocessing by the Gegenbauer reconstruction method. Neuroimage 20:489–502
Wakana S, Jiang H, Nagae-Poetscher LM, van Zijl PC, Mori S (2004) Fiber tract-based atlas of human white matter anatomy. Radiology 230:77–87
Author information
Authors and Affiliations
Contributions
PRK: conceptualization, methodology, and writing original draft preparation. RKJ: visualization, writing—review, and supervision. AK: validation and investigation. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest in publishing the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, P.R., Jha, R.K. & Katti, A. Brain tissue segmentation in neurosurgery: a systematic analysis for quantitative tractography approaches. Acta Neurol Belg 124, 1–15 (2024). https://doi.org/10.1007/s13760-023-02170-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13760-023-02170-9