Abstract
Neural necrosis virus (NNV) of family Nodaviridae affect wide range of fish species with viral encephalopathy and retinopathy causing mass mortality up to 100%. Currently there is no effective treatment and depopulation is only suggested recommendation. New avenues and approach are required to control this harmful malady. In this study we developed an epitope-based vaccine (EBV), against NNV using computation approach. We have selected two conserved proteins RNA-dependent RNA polymerase (RdRP) and capsid proteins. Based on more than ~ 1000 epitopes we selected six antigenic epitopes. These were conjugated to adjuvant and linker peptides to generate a full-length vaccine candidate. Biochemical structural properties were analyzed by Phyre2 server. ProtParam, Molprobity. Ramachandran plot results indicate that 98.7% residues are in a favorable region and 93.4% residues in the favored region. The engineered EBV binds to toll like receptor-5 (TLR5) an important elicitor of immune response. Further molecular docking by PatchDock server reveals the atomic contact energy (i.e. − 267.08) for the best docked model of EBV and TLR5 receptor. The molecular simulation results suggest a stable interaction; the RMSD and RMSF values are 1–4 Ǻ and 1–12Ǻ, respectively. Further we have suggested the best possible codon optimized sequence for its cloning and subsequent purification of the protein. Overall, this is a first report to suggest an in-silico method for generation of an EBV candidate against NNV. We surmise that the method and approach suggested could be used as a promising cure for NNVs.
Similar content being viewed by others
1 Introduction
Mass mortality of commercial fish species like Groupers, Carps, and Tilapia, etc., is a major problem of aquaculture industry (Fukuda et al. 1996). Groupers (Epinephelus and Mycteroperca) and Sea bass (Lateolabrax japonicus) in hatcheries commonly affected by a deadly viral pathogen known as neural necrosis virus (Oh et al. 2006). Neural or nervous necrosis virus also known as Betanodavirus, belongs to family of Nodaviridae. It causes severe viral nervous necrosis (VNN) or viral encephalopathy and retinopathy (VER) in fishes(Yamashita et al. 2005).The infection was accounted initially during the years 1989–1991 when it was related with high mortalities in fingerlings of marine origin and was depicted at first as a picornavirus-like infection. During the same time it was also reported from Australia, Norway, France and Japan (Glazebrook et al. 1990; Mori et al. 1992). Betanodaviruses are non-enveloped, spherical, single-stranded positive sense RNA viruses. The average viral capsid diameter is 30–37 nm. The viral genome is partitioned into two single-stranded RNA molecules RNA-1 (3.1 Kb) and RNA2 (1.2 Kb). A subgenomic transcript, known as RNA3 (0.4 Kb), is synthesized during RNA replication from the 3′ terminus of RNA-1 (Jia et al. 2015; Mori et al. 1992). The crystal structure of a Grouper nervous necrosis virus (GNNV) has been experimentally analyzed by X-ray crystallography. It consists of 180 Capsid subunits, and each capsid protein exhibits three significant areas: (1) the N-terminal arm, an inter subunit expansion at the internal surface; (2) the shell space (S-area), a jelly-like structure; and (3) protrusion area (P-area) framed by trifold trimeric projections (Chen et al. 2015). Betanodavirus affects fingerlings and larval stages of more than 40 species of Teleosts (Nakai et al. 2009). Viral RNA entry in to host neuronal cytoplasm is assisted by coat protein and its interactions with neural receptor domains. Within the nerve cells (Brain and Retina), the viral RNA-1 is translated to produce RNA dependent RNA polymerase (RdRp). RdRp along with other accessory proteins in the cytoplasm makes the viral factories which synthesize dsRNA from the ssRNA (+). The dsRNA is transcribed and replicates to produce new mRNA and ssRNA (+) genomes. The virus assembles with its genomic RNA and capsid protein and through lytic cycle new virions are released (Bandin and Souto 2020; Krishnan et al. 2020a, b).
Vaccine development can be promising for promoting immunity among commercial fish species. Till now no proper therapeutic approach or regimens has been explored to remove such deadly viruses from aquaculture ecosystems (Hazreen-Nita et al. 2019). The only problem is long hit and trials for predicting better vaccine candidates (Kaushik 2019; Michel-Todó et al. 2020). In-silico methods can assist in reducing the effort in screening best putative epitope-based vaccine candidates. The approach involves online web servers and tools, and high-throughput algorithms (like artificial neural networks, Hidden Markov models, and artificial intelligence). Mucosal and Neural immunology of fin fish and shell fish can be supported using epitope-based peptide vaccines (Das and Salinas 2020). Nasopharyngeal mucosal associated lymphoid in Teleost fishes possess large number of leucocytes at intraepithelium and lamina propria sites, and also nasal vaccines were found successful in rainbow trout species (LaPatra et al. 2015). Recent molecular studies of NNV by RNA sequencing suggested that coding sequences related to collagen synthesis and adhesion molecules were increased in the persistent (infected) stage, may suppress the fish immune responses upon NNV infection (Ge et al. 2020). Nanoparticle based lateral flow biosensors were also designed in recent studies to identify NNV easily in fingerlings of mariculture importance (Toubanaki et al. 2020). In this study we targeted proteins of betanodavirus to screen epitopes that have high antigenicity and can interact with MHC like molecules in fishes. We crafted the full-fledged vaccine structure and found its interaction with Toll like receptor-5 (TLR-5) by deploying advanced molecular docking and simulation analysis. Flagellin protein used as adjuvant molecule for vaccine designing as it showed interaction with TLR5 (Tsujita et al. 2006). As in recent studies it was found mass mortality of fresh water fish Tilapia (Oreochromis niloticus) in Egypt due to NNV, this study will be useful for designing better future treatments (Taha et al. 2020). It clearly indicates this problem is associated with marine as well as freshwater aquaculture. NNV have different strains Barfin flounder nervous necrosis virus (BFNNV), Red-spotted grouper nervous necrosis virus (RGNNV), Striped Jack nervous necrosis virus (SJNNV), Tiger puffer nervous necrosis virus (TPNNV), and Seven banded grouper nervous necrosis virus (SBGNNV). We considered SBGNNV as it was less studied but imparts harmful impact on fisheries production (Zorriehzahra 2020). Currently India stand second in world fish production, estimated as 12 million metric tons (MMT) while China is in first position with more than 60MMT fisheries production, to achieve disease free greater production targets is still primary need of world as it is useful for maintaining food security among people(DAHD&F 2018–2019). Latest computational technologies are associated with multiple domains including Life sciences, Environmental sciences, Drug designing, and Vaccine designing. Many algorithm based in-silico approaches are designed to analyze biological data. In one of the recent studies it was found MapReduce oriented architecture (Kamal et al. 2016) that uses several algorithms for management and screening of biological data (like nucleotide and protein sequences). Similarly now a day’s modern sensor technology, based on artificial intelligence and GPRS are already available for environmental monitoring and such technologies saves user time as well as money (Liu 2020). Such advancements of computer sciences in interdisciplinary fields make research more efficient, in a similar way our study is also very unique and shows in-depth application of computational biology in accessing pathogenic proteomic information and assisted us in designing peptide based vaccine against them. Latest in-silico vaccine developments assisted scientific community in identifying epitope-based vaccines against various human pathogens including SARS-CoV2 (Joshi et al. 2020; Akhtar et al. 2020), Tropheryma whipplei bacteria (Joshi et al. 2020), Dengue virus (Krishnan et al. 2020a, b; Krishnan et al. 2021), and Candida aurius fungus (Akhtar et al. 2020). Such latest studies provide motivational support and opens doors to conduct research on other animal groups including fish species and its pathogens. Our study provides a cheapest and alternative approach of crafting vaccine against such harmful pathogens responsible for pulling down fisheries production. In this study we designed epitope-based vaccine against NNV affecting fish species by deploying latest in-silico peptide screening tools, molecular docking servers and molecular dynamic simulation strategies. Here we have also designed complete vaccine model by associating adjuvants and finalized epitopes, for this, modern bioinformatics algorithms based on machine learning approach. In this study we have also introduced in-silico cloning for final vaccine construct which holds future scope for its easy wet-lab synthesis and further testing on animal model.
2 Methodology
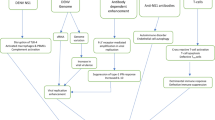
Architectural flow chart is being given in Fig. 1.
2.1 Protein selection and allergenicity prediction
We conducted proteomic data screening SBGNNV using ViPR (Pickett et al. 2012), IEDB and NCBI-Genbank databases. AllergenFP (Dimitrov et al. 2014) was used to calculate Tanimoto similarity index of allergenicity prediction that was found to be useful in finalizing viral proteins.
2.2 Prediction of epitopes and antigenicity
NetCTL 1.2 server (Larsen et al. 2007) was used to detect epitopes of selected proteins; it is based on artificial neural network (ANN) algorithms. It assisted us in determining MHC Class-I supertypes (total 12, namely A1, A2, A3, A24, A26, B7, B8, B27, B39,B44,B58, B62) interacting with epitopes, (small peptides of 9 residues).Threshold was set ≥ 1.2 for cumulative score based on proteasomal C-terminal cleavage and Transporter associated with antigen processing (TAP).VaxiJenserver (Doytchinova and Flower 2007) was used to predict antigenicity of the selected peptide, the threshold value was set as ≥ 0.4.
2.3 Vaccine engineering and structural prediction
The N and C terminal of flagellin protein, RS09 peptide were used as adjuvants (Negahdaripour et al. 2017; Yazdani et al. 2020). Pan DR epitope (PADRE) sequence was used to counter the problems caused by polymorphic HLA alleles and to improve the stability of the vaccine (Hasan et al. 2019). To link the epitopes and adjuvants linker sequence GGS was used (Yazdani et al. 2020). After the construction of vaccine, its physiochemical parameters were evaluated using ExPASy ProtParam tool. This server helps to predict the stability index, grand average of hydropathy (GRAVY) score, Isoelectric point, and aliphatic index (Gasteiger et al. 2005).Ramachandran plot was constructed using Molprobity server (Davis et al. 2007). Structural validations of full-fledged vaccine structure were predicted by deploying Phyre2 (Kelley et al. 2015), it is based on advanced CASP (critical assessment of protein structure prediction) and homology modeling methods.
2.4 Molecular docking and simulation studies
ZDock (Pierce et al. 2014) and PatchDock (Schneidman-Duhovny et al. 2005) web servers were used for docking studies and to analyze best molecular interactions of TLR5 and full-fledged vaccine. Molecular simulation studies were performed by deploying Schrodinger Academic Desmond MDS suite (Bowers et al. 2006).
2.5 In-silico cloning
J-CAT server (Grote et al. 2005) was used to determine coding sequence of predicted vaccine, along with codon adaptive index. It was further use to design recombinant plasmid by deploying SnapGene tool (from GLS Biotech, available at snapgene.com), that can be used for cloning and producing vaccine and develops more practical insights of epitope-based peptide vaccine (Chaudhuri et al. 2020).
3 Results
3.1 Proteomic screening and epitope selection
After analyzing proteomic data two proteins RNA-dependent RNA polymerase and Coat Protein of Betanodavirus (SBGNNV) were selected, as they were found non-allergen (Table 1). NetCTL 1.2 server results 974 epitopes for RNA-Dependent RNA polymerase and 330 epitopes for Coat protein, that were analyzed on the basis of their best overall interaction score to MHC Supertypes A1, B27, and A3 (also mention from 12 MHC types). This interaction score is cumulative score of proteasomal processing, TAP transport efficiency and epitope affinity towards MHC supertypes. Figure 2 indicates Plot of cumulative score for epitopes considered for further analysis. Then the screened epitopes were subjected towards VaxiJen server for immunogenicity or antigenicity prediction, that result in to total of 6 putative epitopes out of them 2 were from RNA-dependent RNA polymerase and 4 were from coat protein (Table 2). ProtParam tool was used to analyze physiochemical properties of selected Epitopes (Table 3).
3.2 Vaccine engineering and structural analysis
Epitopes selected in previous section were then used to craft full-fledged vaccine by adding adjuvant, and linkers. Three adjuvants RS09 (APPHALS), PADRE sequence (AKFVAAWTLKAAA) and N-terminal and C-terminal sequence of Salmonella typhimurium flagellin protein were linked to the epitopes with the help of GGS linker. RS09 adjuvant is a synthetic Toll like receptor-4 agonist and flagellin is a Toll like receptor-5 agonist. Both are involved in the activation of innate and adaptive immune response (Shanmugam et al. 2012).
Toll like receptors are involved in the activation of innate immunity and also improve antigen presentation by Antigen Presenting Cells (APCs). PADRE sequence provides stability to the vaccine and it has also been used as adjuvant in vaccine design (Hasan et al. 2019). PsiPred server (McGuffin et al. 2000) was used to find out coils, Beta sheets and Helices in the vaccine designed (Fig. 3).The rectangular boxes shown in the (Fig. 3) are the selected or finalized epitopes used in vaccine designing (Table 3). Phyre 2 server assisted in developing best 3D homology model for the vaccine crafted, shown in Fig. 4, epitope-based vaccine determined have 395 residues and pink color indicates helices, while yellow color indicates strands in given representation.
TLR5 receptor (PDBID: 3J0A) structure was obtained from rcsb-Pdb database (https://www.rcsb.org/) and its Chain A was selected for Molecular docking and Simulation analysis with the conjugated vaccine. Ramachandran plot for predicted vaccine indicates 93.4% of all residues in favored region and 98.7% of all residues were in allowed region (Fig. 5). This clearly indicates structural stability of epitope based vaccine (EBV) (Fig. 5).
3.3 Molecular docking and simulation studies
Docking between EBV and TLR05 receptor was performed by Zdock and PatchDock-Firedock servers. PatchDock utilized transformation parameters or coordinates centered x, y, and z as 1.35, 0.54, 0.88 respectively; and size frame as 58.83, 33.25, and 68.80 respectively. For best model out of 1000 docked structure atomic contact energy (ACE) score was obtained as − 267.08. In Fig. 6 clearly shows the interaction of EBV (Pink) and TLR5receptor (Blue) was perfect. Hydrogen bond interactions between EBV residues to TLR5 residues are as follows: Gln129 to Leu 208 (3.1 Ǻ), Val 161 to Asp 101(3.4 Ǻ), Ala 293 to Leu 343 (2.5 Ǻ), Ser73 to Asp 433 (3.1 Ǻ), and Asn 142 to Cys24 (2.5 Ǻ).
Molecular simulation study was performed by deploying Schrodinger Academic Desmond MDS suite (Bowers et al. 2006). The side chain protonation state of amino acids in the TLR5 and EBV were assigned in relation to the pH 7.0 through the pKa prediction by the propka tool. The resulting structure was optimized and then energy minimized. The resulting complex was then solvated in TIP3P water model with orthorhombic system shape and ten angstroms periodic boundary conditions (PBC). Overall charge of the system was set to neutral through addition of appropriate Na+ and Cl− counter ions. Physiological salt concentration of 0.15 Molar was achieved through addition of NaCl. The resulting solvated system was once again energy minimized using the Desmond Minimization. The default relaxation protocol was used for preparatory phase MDS. The extent of the production molecular dynamics (MD) was 10 ns and the conformations were recorded every 10picoseconds under OPLS_2005 force field. Root mean square density and root mean square fluctuation plots found under the selectable range (Fig. 7), i.e., 1–4 Ǻ for RMSD values and 1–12 Ǻ for RMSF values. This clearly indicates that selected docked complex was found to be most stable, and EBV can be further used for in-silico cloning studies to know expression state.
Molecular simulation analysis of docked complex: A the RMSD density distribution histogram for the docked complex TLR5 and vaccine adjuvant are shown. B The RMSF plots of the docked complex TLR5 and vaccine adjuvant are shown. Least RMS fluctuation can be seen in these interface residues, that acts as indicator of vaccine interactional stability
3.4 EBV codon adaptation and in-silico cloning
Codon usage adaptation is significant for enhancing the expression of the EBV in prokaryotic hosts (Escherichia coli strain K12 was selected). After the codon adaptation of the vaccine, the Codon Adaptation Index was predicted to be 1 which ensures high degree of the sequence expression and the GC content of the sequence were found to be 52% in J-CAT server (Fig. 8). In-silico cloning of the vaccine was conducted by deploying SnapGene restriction cloning module in SnapGenetool. The final EBV gene was inserted within the region flanks between 1000 and 2000 base pairs as ORF2 resides here and provides ease in insertion with multiple restriction sites (Fig. 9).
4 Discussion
Recent attempts for epitope-based vaccine crafting against viral pathogens of fin fishes and shell fishes were found relatively successful as compared to web lab experiments (Momtaz et al. 2019). Whole concept about EBVs is itself very novel and easy in application for everyone, in recent studies it was found successful in determining epitope-based vaccine candidates for SARS-CoV2, Dengue virus, and even for rare bacterium like Tropheryma whipplei affecting humans (Joshi et al. 2020; Krishnan et al. 2020a, b).For chickens, shrimps epitope-based vaccines were already found effective and it is required to design such vaccines against fin fishes also (Tan et al. 2016). Fish pathogens Edwardsiella tarda and Flavobacterium columnare was successfully targeted by computer-aided epitope-based vaccine candidates (Mahendran et al. 2016). T-cell epitopes can induce primary immune response in the hosts as they have good interactional activity with MHC Class-I molecules (Brusic et al. 2004). In present study we successfully found six epitopes by deploying RNA dependent RNA polymerase and Coat proteins of NNV (neural necrosis virus) that mostly affect Groupers (Epinephelus spp.), Gray mullet (Mugil cephalus), Rainbow trout (Onchorynchus mykiss) and Sea bass (Lates calcarifer) fish species. This virus was not only imparting harmful effect on physiology of fish species but also has capacity to decline global fish production, like in India it from two decades shrimp and fin fish industry suffered (John et al. 2014).The EBV modeling was very first time deployed in this study as in earlier studies only few HLA alleles of MHC Class I was considered (Mahendran et al. 2016). We tried to show a better designing of vaccine by adding adjuvant and linkers. Also, the structural stability of EBV was successfully determined by Ramachandran plot analysis, this plot assisted in analyzing residues preference present on secondary structures in EBV. Molecular docking and simulation studies reveals perfect model of EBV with TLR-5 receptor, docking shows EBV interaction with binding pocket of TLR5 receptor. TLR5 in Teleosts have tendency of attraction towards flagellin protein also, so we used flagellin head and tail residual peptides in EBV crafting, it was found to have role in generating inflammatory response to develop innate immunity in fishes (Palti 2011; Gao et al. 2018). RMSD and RMSF plots have greater significance in determining stability in docked models for a long run (Martínez 2015), here we used best simulation tool Desmond to access stable interactions of EBV with TLR5 receptor (on a standard benchmark, Desmond's selection over a conventional tools is its regular updating, high performance, and user-friendly). In-silico cloning based on codon usage adaptation was used as this approach provides computer based assessment of cloning of genes encoding vaccine candidates (Ullah et al. 2020), here we deployed this same concept to find out coding sequences that encodes EBV and then subjected such sequences to determine codon usage pattern and lastly for plasmid insertion. In fish vaccine designing this method was first time deployed. This method is not only new but also open many dimensions for associating fish immunology with bioinformatics. This approach is efficient and effective, allows easy wet-lab discoveries.
5 Conclusion
EBV was found against betanodavirus or nervous necrosis virus that mostly affect Grouper, and Sea bass fish species. To enhance Aquaculture production disease elimination and new regimens are always required. In this study EBV passed all the computational (biochemical structural and functional) assessments. Most stable hexaepitope vaccine with flagellin like adjuvants makes it more effective regimen for disease control. RdRp and Coat protein can easily targeted by fish immune systems after wet-lab validations. This new method will save time and can be used to predict epitope-based vaccines for other harmful pathogens affecting fish species.
Abbreviations
- AI:
-
Artificial intelligence
- ANN:
-
Artificial neural networks
- HMM:
-
Hidden Markov model
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
References
Akhtar N, Joshi A, Singh B, Kaushik V (2020) Immuno-informatics quest against COVID-19/SARS-COV-2: determining putative T-Cell epitopes for vaccine prediction. Infect Disord Drug Targets
Akhtar N, Joshi A, Kaushik V, Kumar M, Mannan MA (2021) In-silico design of a multivalent epitope-based vaccine against Candida auris. Microb Pathog 155:104879
Bandín I, Souto S (2020) Betanodavirus and VER disease: a 30-year research review. Pathogens 9(2):106
Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: SC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. IEEE, p 43-43
Brusic V, Bajic VB, Petrovsky N (2004) Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods 34(4):436–443
Chaudhuri D, Datta J, Majumder S, Giri K (2020) In silico designing of peptide based vaccine for hepatitis viruses using reverse vaccinology approach. Infect Genet Evol 84:104388
Chen NC, Yoshimura M, Guan HH, Wang TY, Misumi Y, Lin CC et al (2015) Crystal structures of a piscine betanodavirus: mechanisms of capsid assembly and viral infection. PLoSPathog 11(10):e1005203
DAHD&F (2018–2019) DAHD&F Annual report—2018–2019. Department of animal husbandry, dairying and fisheries, ministry of agriculture. Government of India (2019). http://www.dahd.nic.in/documents/reports. Accessed 22 Jun 2020
Das PK, Salinas I (2020) Fish nasal immunity: From mucosal vaccines to neuroimmunology. Fish Shellfish Immunol 104:165–171
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X et al (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35(suppl_2):W375–W383
Dimitrov I, Naneva L, Doytchinova I, Bangov I (2014) AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics 30(6):846–851
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8(1):4
Fukuda Y, Furuhashi M, Nakai T (1996) Mass mortality of cultured sevenband grouper, Epinephelusseptemfasciatus, associated with viral nervous necrosis. Fish Pathol 31(3):165–170
Gao Q, Yue Y, Min M, Peng S, Shi Z, Sheng W, Zhang T (2018) Characterization of TLR5 and TLR9 from silver pomfret (Pampus argenteus) and expression profiling in response to bacterial components. Fish Shellfish Immunol 80:241–249
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. p 571–607
Ge H, Lin K, Zhou C, Lin Q, Zhang Z, Wu J et al (2020) A multi-omic analysis of orange-spotted grouper larvae infected with nervous necrosis virus identifies increased adhesion molecules and collagen synthesis in the persistent state. Fish Shellfish Immunol 98:595–604
Glazebrook JS, Heasman MP, De Beer SW (1990) Picorna-like viral particles associated with mass mortalities in larval barramundi, Lates calcarifer Bloch. J Fish Dis 13(3):245–249
Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D (2005) JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33(suppl_2):W526–W531
Hasan M, Islam S, Chakraborty S, Mustafa AH, Azim KF, Joy ZF, Hossain MN, Foysal SH, Hasan MN (2019) Contriving a chimeric polyvalent vaccine to prevent infections caused by herpes simplex virus (type-1 and type-2): an exploratory immunoinformatic approach. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2019.1647286
Hazreen-Nita M, Azila A, Mukai Y, Firdaus-Nawi M, Nur-Nazifah M (2019) A review of betanodavirus vaccination as preventive strategy to viral nervous necrosis (VNN) disease in grouper. Aquacult Int 27(5):1565–1577
Jia P, Jia KT, Yi MS (2015) Complete genome sequence of a fish nervous necrosis virus isolated from sea perch (Lateolabrax japonicus) in China. Genome Announc 3(3)
John KR, George MR, Jeyatha B, Saravanakumar R, Sundar P, Jithendran KP, Koppang EO (2014) Isolation and characterization of Indian betanodavirus strain from infected farm-reared Asian seabass Lates calcarifer (Bloch, 1790) juveniles. Aquac Res 45:1481–1488
Joshi A, Kaushik V (2021) In-silico proteomic exploratory quest: crafting T-cell epitope vaccine against Whipple’s disease. Int J Pept Res Ther 27:169–179
Joshi A, Joshi BC, Mannan MAU, Kaushik V (2020) Epitope based vaccine prediction for SARS-COV-2 by deploying immuno-informatics approach. Inf Med Unlocked 19:100338
Kamal S, Ripon SH, Dey N, Ashour AS, Santhi V (2016) A MapReduce approach to diminish imbalance parameters for big deoxyribonucleic acid dataset. Comput Methods Programs Biomed 131:191–206
Kaushik V (2019) In Silico identification of epitope-based peptide vaccine for Nipah virus. Int J Pept Res Ther 1-7
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Krishnan S, Joshi A, Kaushik V (2020b) T cell epitope designing for dengue peptide vaccine using docking and molecular simulation studies. Mol Simul 46(10):787–795
Krishnan R, Kim JO, Qadiri SSN, Kim JO, Oh MJ (2020a) Early viral uptake and host-associated immune response in the tissues of seven-band grouper following a bath challenge with nervous necrosis virus. Fish Shellfish Immunol 103:454–463
Krishnan S, Joshi A, Akhtar N, Kaushik V (2021) Immunoinformatics designed T cell multi epitope dengue peptide vaccine derived from non structural proteome. Microb Pathog 150:104728
LaPatra S, Kao S, Erhardt EB, Salinas I (2015) Evaluation of dual nasal delivery of infectious hematopoietic necrosis virus and enteric red mouth vaccines in rainbow trout (Oncorhynchus mykiss). Vaccine 33(6):771–776
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M (2007) Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform 8(1):424
Liu Q (2020) Intelligent environmental monitoring system based on multi-sensor data technology. Int J Ambient Comput Intell 11(4):57–71
Mahendran R, Jeyabaskar S, Sitharaman G, Michael RD, Paul AV (2016) Computer-aided vaccine designing approach against fish pathogens Edwardsiellatarda and Flavobacterium columnare using bioinformatics softwares. Drug Des Dev Ther 10:1703
Martínez L (2015) Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 10(3):e0119264
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405
Michel-Todó L, Bigey P, Reche PA, Pinazo MJ, Gascón J, Alonso-Padilla J (2020) Design of an epitope-based vaccine ensemble for animal trypanosomiasis by computational methods. Vaccines 8(1):130
Momtaz F, Foysal J, Rahman M, Fotedar R (2019) Design of epitope based vaccine against shrimp white spot syndrome virus (WSSV) by targeting the envelope proteins: an immunoinformatic approach. Turk J Fish Aquat Sci 19(2):59–69
Mori KI, Nakai T, Muroga K, Arimoto M, Mushiake K, Furusawa I (1992) Properties of a new virus belonging to Nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology 187(1):368–371
Nakai T, Sugaya T, Nishioka T, Mushiake K, Yamashita H (2009) Current knowledge on viral nervous necrosis (VNN) and its causative betanodaviruses. Isr J Aquac Bamidgeh 61(1–4):198–207
Negahdaripour M, Eslami M, Nezafat N, Hajighahramani N, Ghoshoon MB, Shoolian E, Dehshahri A, Erfani N, Morowvat MH, Ghasemi Y (2017) A novel HPV prophylactic peptide vaccine, designed by immunoinformatics and structural vaccinology approaches. Infect Genet Evol 54:402–416. https://doi.org/10.1016/j.meegid.2017.08.002
Oh MJ, Jung SJ, Kitamura SI, Kim HY, Kang SY (2006) Viral diseases of olive flounder in Korean hatcheries. J Ocean Univ China 5(1):45–48
Palti Y (2011) Toll-like receptors in bony fish: from genomics to function. Dev Comp Immunol 35(12):1263–1272
Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V et al (2012) ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res 40(D1):D593–D598
Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z (2014) ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 30(12):1771–1773
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33(suppl_2):W363–W367
Shanmugam A, Rajoria S, George AL, Mittelman A, Suriano R, Tiwari RK (2012) Synthetic toll like Receptor-4 (TLR-4) agonist peptides as a novel class of adjuvants. PLoS ONE 7:e30839. https://doi.org/10.1371/journal.pone.0030839
Taha E, Shawky M, Ahmed B, Moustafa M, Yousif A, Abdelaziz M (2020) Emergence of viral nervous necrosis is associated with mass mortality in hatchery-reared tilapia (Oreochromis niloticus) in Egypt. Aquac Int 28:1811–1823
Tan L, Zhang Y, Liu F, Yuan Y, Zhan Y, Sun Y et al (2016) Infectious bronchitis virus poly-epitope-based vaccine protects chickens from acute infection. Vaccine 34(44):5209–5216
Toubanaki DK, Margaroni M, Prapas A, Karagouni E (2020) Development of a nanoparticle-based lateral flow strip biosensor for visual detection of whole nervous necrosis virus particles. Sci Rep 10(1):1–12
Tsujita T, Ishii A, Tsukada H, Matsumoto M, Che FS, Seya T (2006) Fish soluble Toll-like receptor (TLR) 5 amplifies human TLR5 response via physical binding to flagellin. Vaccine 24(12):2193–2199
Ullah MA, Sarkar B, Islam SS (2020) Exploiting the reverse vaccinology approach to design novel subunit vaccines against Ebola virus. Immunobiology 225(3):151949
Yamashita H, Fujita Y, Kawakami H, Nakai T (2005) The efficacy of inactivated virus vaccine against viral nervous necrosis (VNN). Fish Pathol 40(1):15–21
Yazdani Z, Rafiei A, Valadan R, Ashrafi H, Pasandi MS, Kardan M (2020) Designing a potent L1 protein-based HPV peptide vaccine: a bioinformatics approach. Comput Biol Chem. https://doi.org/10.1016/j.compbiolchem.2020.107209
Zorriehzahra MJ (2020) Viral nervous necrosis disease. Emerging and reemerging viral pathogens. Academic Press, New York, pp 673–703
Acknowledgements
The authors would like to acknowledge School of Bioengineering and Biosciences for the infrastructure support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of the interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Joshi, A., Pathak, D.C., Mannan, M.Au. et al. In-silico designing of epitope-based vaccine against the seven banded grouper nervous necrosis virus affecting fish species. Netw Model Anal Health Inform Bioinforma 10, 37 (2021). https://doi.org/10.1007/s13721-021-00315-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-021-00315-5