Abstract
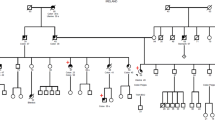
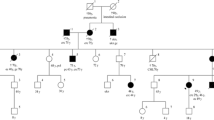
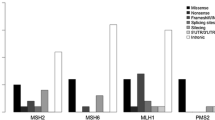
Immunohistochemistry of mismatch repair proteins is a universal strategy for Lynch syndrome screening. In this case, Lynch syndrome was suspected, because MLH1 and PMS2 expression was negative by IHC. However, mismatch repair genetic analysis revealed a variant of unknown significance of c.454-13A > G in MLH1. Therefore, we performed reverse transcription-PCR using mRNA extracted from the patient’s lymphocytes and detected a heterozygous gene allele indicating splicing abnormalities that complex splicing, with exon 5 followed by only the first codon (ACG) of exon 6 and leading to exon 7 of the MLH1. Two years later, this mutation was corrected to “likely pathogenic”. For Lynch syndrome in which mismatch repair protein expression is undetectable by immunohistochemistry, reverse transcription-PCR may be useful to identify an intronic variant of unknown significance as the likely pathogenic variant.





Similar content being viewed by others
References
Dominguez-Valentin M, Sampson JR, Seppälä TT et al (2020) Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the prospective Lynch syndrome database. Genet Med 22:15–25
Hampel H, Frankel WL, Martin E et al (2005) Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352:1851–1860
Tamura K, Kaneda M, Futagawa M et al (2019) Genetic and genomic basis of the mismatch repair system involved in Lynch syndrome. Int J Clin Oncol 24:999–1011
Piñero TA, Soukarieh O, Rolain M et al (2020) MLH1 intronic variants mapping to + 5 position of splice donor sites lead to deleterious effects on RNA splicing. Fam Cancer. https://doi.org/10.1007/s10689-020-00182-5
Takahashi M, Furukawa Y, Shimodaira H et al (2012) Aberrant splicing caused by a MLH1 splice donor site mutation found in a young Japanese patient with Lynch syndrome. Fam Cancer 11:559–564
Vasen HF, Watson P, Mecklin JP et al (1999) New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116:1453–1456
Snowsill T, Coelho H, Huxley N et al (2017) Molecular testing for Lynch syndrome in people with colorectal cancer: systematic reviews and economic evaluation. Health Technol Assess 21:1–238
Paredes SR, Chan C, Rickard MX (2020) Immunohistochemistry in screening for heritable colorectal cancer: what to do with an abnormal result. ANZ J Surg 90:702–707
Kohonen-Corish M, Ross VL, Doe WF et al (1996) RNA-based mutation screening in hereditary nonpolyposis colorectal cancer. Am J Hum Genet 59:818–824
Peltomäki P (2016) Update on Lynch syndrome genomics. Fam Cancer 15:385–393
Zhang MQ (1998) Statistical features of human exons and their flanking regions. Hum Mol Genet 7:919–932
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Nishikubo, T., Masui, K., Koyama, F. et al. Immunohistochemistry and RNA-sequencing have been useful in evaluating the pathological significance of a non-consensus site intronic variant in suspected cases of Lynch syndrome. Int Canc Conf J 10, 186–190 (2021). https://doi.org/10.1007/s13691-021-00474-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13691-021-00474-2