Abstract
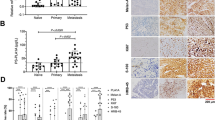
Existing clinical indicators for metastatic risk classification and patient treatment of uveal melanoma (UM) in the Asian population are limited. Preferentially expressed antigen in melanoma (PRAME) has gained attention in the prognosis of cancers and considered as a potential biomarker in many tumors including UM. Therefore, this study investigated the expression of PRAME and its association with loss of nuclear BAP1 (nBAP1) as well as its correlation with clinicopathological parameters and patient outcome. Immunohistochemical expression of PRAME and BAP1 proteins were assessed in 66 prospective cases of UM. mRNA expression level was measured by quantitative real-time PCR. Kaplan–Meier curves and Cox proportional hazard models were used to analyze the correlation of protein expression with clinicopathological parameters, metastasis-free survival and overall survival. Nuclear PRAME (nPRAME) expression and loss of nBAP1 were observed in 24 and 62% cases, respectively. PRAME mRNA expression level was found to be upregulated in 64% (7/11) of metastatic patients. mRNA and immunoexpression of nPRAME were statistically significant with many clinicopathological high-risk factors. On univariate and multivariate analyses, high mitotic activity, extraocular invasion and presence of nPRAME expression were statistically significant (p < 0.05). On Kaplan–Meier survival analysis, patients expressing PRAME had significantly reduced metastasis-free survival (MFS) and overall survival (OS). MFS and OS were also reduced in patients expressing PRAME along with loss of nBAP1. Our data show that nPRAME expression, in combination with loss of nBAP1, could be a useful predictive biomarker in the therapeutic management of UM patients at high risk.



Similar content being viewed by others
References
Lee WS, Lee J, Choi JJ, Kang HG, Lee SC, Kim JH. Paired comparisons of mutational profiles before and after brachytherapy in asian uveal melanoma patients. Sci Rep. 2021;11(1):18594. https://doi.org/10.1038/s41598-021-98084-8.
Branisteanu DC, Bogdanici CM, Branisteanu DE, Maranduca MA, Zemba M, Balta F, et al. Uveal melanoma diagnosis and current treatment options (Review). Exp Ther Med. 2021;22(6):1428. https://doi.org/10.3892/etm.2021.10863.
Jager MJ, Shields CL, Cebulla CM, Abdel-Rahman MH, Grossniklaus HE, Stern MH, et al. Uveal melanoma. Nat Rev Dis Primers. 2020;6(1):24. https://doi.org/10.1038/s41572-020-0158-0.
Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64(20):7205–9. https://doi.org/10.1158/0008-5472.Can-04-1750.
Harbour JW. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Biol. 2014;1102:427–40. https://doi.org/10.1007/978-1-62703-727-3_22.
van Essen TH, van Pelt SI, Versluis M, Bronkhorst IH, van Duinen SG, Marinkovic M, et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol. 2014;98(12):1738–43. https://doi.org/10.1136/bjophthalmol-2014-305047.
Aaberg TM, Covington KR, Tsai T, Shildkrot Y, Plasseraud KM, Alsina KM, et al. Gene expression profiling in uveal melanoma: five-year prospective outcomes and meta-analysis. Ocul Oncol Pathol. 2020;6(5):360–7. https://doi.org/10.1159/000508382.
Field MG, Decatur CL, Kurtenbach S, Gezgin G, van der Velden PA, Jager MJ, et al. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin Cancer Res. 2016;22(5):1234–42. https://doi.org/10.1158/1078-0432.Ccr-15-2071.
Lezcano C, Jungbluth AA, Nehal KS, Hollmann TJ, Busam KJ. PRAME expression in melanocytic tumors. Am J Surg Pathol. 2018;42(11):1456–65. https://doi.org/10.1097/pas.0000000000001134.
Szczepanski MJ, DeLeo AB, Łuczak M, Molinska-Glura M, Misiak J, Szarzynska B, et al. PRAME expression in head and neck cancer correlates with markers of poor prognosis and might help in selecting candidates for retinoid chemoprevention in pre-malignant lesions. Oral Oncol. 2013;49(2):144–51. https://doi.org/10.1016/j.oraloncology.2012.08.005.
Thongprasert S, Yang PC, Lee JS, Soo R, Gruselle O, Myo A, et al. The prevalence of expression of MAGE-A3 and PRAME tumor antigens in East and South East Asian non-small cell lung cancer patients. Lung Cancer. 2016;101:137–44. https://doi.org/10.1016/j.lungcan.2016.09.006.
Epping MT, Hart AA, Glas AM, Krijgsman O, Bernards R. PRAME expression and clinical outcome of breast cancer. Br J Cancer. 2008;99(3):398–403. https://doi.org/10.1038/sj.bjc.6604494.
Lezcano C, Pulitzer M, Moy AP, Hollmann TJ, Jungbluth AA, Busam KJ. Immunohistochemistry for PRAME in the distinction of nodal nevi from metastatic melanoma. Am J Surg Pathol. 2020;44(4):503–8. https://doi.org/10.1097/pas.0000000000001393.
Toyama A, Siegel L, Nelson AC, Najmuddin M, Bu L, LaRue R, et al. Analyses of molecular and histopathologic features and expression of PRAME by immunohistochemistry in mucosal melanomas. Mod Pathol. 2019;32(12):1727–33. https://doi.org/10.1038/s41379-019-0335-4.
Lohman ME, Steen AJ, Grekin RC, North JP. The utility of PRAME staining in identifying malignant transformation of melanocytic nevi. J Cutan Pathol. 2021;48(7):856–62. https://doi.org/10.1111/cup.13958.
Coupland S, Kalirai H, Taktak A, Eleuteri A, Damato B. Re: Gelmi et al adding the cancer genome atlas chromosome classes to American Joint Committee on Cancer System offers more precise prognostication in uveal melanoma. Ophthalmology. 2022. https://doi.org/10.1016/j.ophtha.2022.02.031.
Kapoor AG, Kaliki S, Vempuluru VS, Jajapuram SD, Ali MH, Mohamed A. (2020) Posterior uveal melanoma in 321 Asian Indian patients: analysis based on the 8th edition of American Joint Committee Cancer classification. Int Ophthalmol. 40(11): 3087-3096 https://doi.org/10.1007/s10792-020-01494-2
Jha J, Singh MK, Singh L, Pushker N, Bajaj MS, Sen S, et al. Expression of BAP1 and ATM proteins: association with AJCC tumor category in uveal melanoma. Ann Diagn Pathol. 2020;44: 151432. https://doi.org/10.1016/j.anndiagpath.2019.151432.
Ahmadian SS, Dryden IJ, Naranjo A, Toland A, Cayrol RA, Born DE, et al. Preferentially expressed antigen in melanoma immunohistochemistry labeling in uveal melanomas. Ocul Oncol Pathol. 2022;8(2):133–40. https://doi.org/10.1159/000524051.
See TR, Stålhammar G, Phillips S, Grossniklaus HE. BAP1 immunoreactivity correlates with gene expression class in uveal melanoma. Ocul Oncol Pathol. 2020;6(2):129–37. https://doi.org/10.1159/000502550.
Kaštelan S, Antunica AG, Oresković LB, Pelčić G, Kasun E, Hat K. Immunotherapy for uveal melanoma - current knowledge and perspectives. Curr Med Chem. 2020;27(8):1350–66. https://doi.org/10.2174/0929867326666190704141444.
Schefler AC, Koca E, Bernicker EH, Correa ZM. Relationship between clinical features, GEP class, and PRAME expression in uveal melanoma. Graefes Arch Clin Exp Ophthalmol. 2019;257(7):1541–5. https://doi.org/10.1007/s00417-019-04335-w.
Cai L, Paez-Escamilla M, Walter SD, Tarlan B, Decatur CL, Perez BM, et al. Gene expression profiling and PRAME status versus tumor-node-metastasis staging for prognostication in uveal melanoma. Am J Ophthalmol. 2018;195:154–60. https://doi.org/10.1016/j.ajo.2018.07.045.
Ikeda H, Lethé B, Lehmann F, van Baren N, Baurain JF, de Smet C, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6(2):199–208. https://doi.org/10.1016/s1074-7613(00)80426-4.
Chang AY, Dao T, Gejman RS, Jarvis CA, Scott A, Dubrovsky L, et al. A therapeutic T cell receptor mimic antibody targets tumor-associated PRAME peptide/HLA-I antigens. J Clin Invest. 2017;127(7):2705–18. https://doi.org/10.1172/jci92335.
Wadelin F, Fulton J, McEwan PA, Spriggs KA, Emsley J, Heery DM. Leucine-rich repeat protein PRAME: expression, potential functions and clinical implications for leukaemia. Mol Cancer. 2010;9:226. https://doi.org/10.1186/1476-4598-9-226.
Baba H, Kanda M, Sawaki K, Umeda S, Miwa T, Shimizu D, et al. PRAME as a potential biomarker for liver metastasis of gastric cancer. Ann Surg Oncol. 2020;27(6):2071–80. https://doi.org/10.1245/s10434-019-07985-6.
Ding K, Wang XM, Fu R, Ruan EB, Liu H, Shao ZH. PRAME gene expression in acute leukemia and its clinical significance. Cancer Biol Med. 2012;9(1):73–6. https://doi.org/10.3969/j.issn.2095-3941.2012.01.013.
Cazzato G, Mangialardi K, Falcicchio G, Colagrande A, Ingravallo G, Arezzo F, et al. Preferentially expressed antigen in melanoma (PRAME) and human malignant melanoma: a retrospective study. Genes. 2022;13(3):545. https://doi.org/10.3390/genes13030545.
Zhang YH, Lu AD, Yang L, Li LD, Chen WM, Long LY, et al. PRAME overexpression predicted good outcome in pediatric B-cell acute lymphoblastic leukemia patients receiving chemotherapy. Leuk Res. 2017;52:43–9. https://doi.org/10.1016/j.leukres.2016.11.005.
Al-Khadairi G, Naik A, Thomas R, Al-Sulaiti B, Rizly S, Decock J. PRAME promotes epithelial-to-mesenchymal transition in triple negative breast cancer. J Transl Med. 2019;17(1):9. https://doi.org/10.1186/s12967-018-1757-3.
Field MG, Durante MA, Decatur CL, Tarlan B, Oelschlager KM, Stone JF, et al. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget. 2016;7(37):59209–19. https://doi.org/10.18632/oncotarget.10962.
Bronkhorst IH, Jager MJ. Uveal melanoma: the inflammatory microenvironment. J Innate Immun. 2012;4(5–6):454–62. https://doi.org/10.1159/000334576.
Mlecnik B, Bindea G, Pagès F, Galon J. Tumor immunosurveillance in human cancers. Cancer Metastasis Rev. 2011;30(1):5–12. https://doi.org/10.1007/s10555-011-9270-7.
Xu Q, Wang C, Yuan X, Feng Z, Han Z. Prognostic value of tumor-infiltrating lymphocytes for patients with head and neck squamous cell carcinoma. Transl Oncol. 2017;10(1):10–6. https://doi.org/10.1016/j.tranon.2016.10.005.
Li JF, Chu YW, Wang GM, Zhu TY, Rong RM, Hou J, et al. The prognostic value of peritumoral regulatory T cells and its correlation with intratumoral cyclooxygenase-2 expression in clear cell renal cell carcinoma. BJU Int. 2009;103(3):399–405. https://doi.org/10.1111/j.1464-410X.2008.08151.x.
Acknowledgements
Nikhil Kumar is thankful to the Indian Council of Medical Research for providing Junior Research Fellowship.
Funding
This research was funded by Indian Council of Medical Research (F. No. 5/4/6/3/OPH/2019-NCD-II).
Author information
Authors and Affiliations
Contributions
MKS and SK were responsible for the conception and design of the work. NK, LS and MKS contributed to the acquisition, analysis, and interpretation of data for the work. NL and RM provided the enucleated specimens. SS and SK helped in reviewing the histopathology slides. All the authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
None.
Ethical approval
Ethical approval was obtained from the Institute’s Ethical Committee, All India Institute of Medical Sciences (Ref. No. IEC-189/05–04-2019).
Informed consent
Written informed consent was obtained from all the patients before participation in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, N., Singh, M.K., Singh, L. et al. Diagnostic utility of immunohistochemistry in concordance with mRNA analysis of PRAME in the stratification of high-risk uveal melanoma patients. Human Cell 36, 342–352 (2023). https://doi.org/10.1007/s13577-022-00808-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00808-z