Abstract
Introduction
Photoaging is the process by which ultraviolet rays gradually induce clinical and histological changes in the skin through the production and organization of biological molecules, such as elastin, which is critical to skin strength and elasticity. After exposure to radiation, elastin may undergo alternative mRNA splicing, resulting in modified proteins that contribute to the formation of aging characteristics, such as solar elastosis. The present work aimed to study two different forms of elastin under these conditions: normal elastin and elastin that had been altered in exon 26A.
Methods
These different forms of elastin were characterized for gene expression by quantitative real-time polymerase chain reaction (qPCR) and for protein expression by immunohistochemistry of ex vivo skins (from photoexposed and non-photoexposed areas) and in vitro reconstituted skin. In addition, up- and downstream molecules in the elastin signaling cascade were evaluated.
Results
As a result, a significant increase in the gene expression of elastin 26A was observed in both ex vivo photoexposed skin tissues and the in vitro photoexposed reconstituted skins. Additionally, significant increases in the gene expression levels of matrix metalloproteinase-12 (MMP12) and lysyl oxidase (LOX) were observed in the ex vivo skin model. The evaluation of protein expression levels of some photoaging markers on the reconstituted skin revealed increased tropoelastin and fibrillin-1 expression after photoexposure.
Conclusion
This work contributes to a better understanding of the biological mechanisms involved in photoaging, making it possible to obtain new strategies for the development of dermocosmetic active ingredients to prevent and treat skin aging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Photoaging is the process by which ultraviolet rays gradually induce clinical and histological changes in the skin through the production and organization of biological molecules, such as elastin, which is critical to skin strength and elasticity. |
After exposure to radiation, elastin may undergo alternative mRNA splicing, resulting in modified proteins that contribute to the formation of aging characteristics, such as solar elastosis. |
We verified that a significant increase in the gene expression of a different form of elastin (elastin 26A) was observed in ex vivo photoexposed skin tissues and in in vitro photoexposed reconstituted skins. |
This work contributes to a better understanding of the biological mechanisms involved in photoaging, making it possible to obtain new strategies for the development of dermocosmetic active ingredients to prevent and treat skin aging. |
Introduction
The skin represents the interface of the human body with the environment and is exposed to several factors such as physical impacts, microorganism invasion, xenobiotic agents, pollution, radiation, and dehydration. This implies that diverse structural and physiological transformations can occur with aging, which can be associated with pathological processes. Therefore, maintenance of cutaneous integrity is extremely important so that the body can defend itself against various aggressions and even prevent skin aging [1].
Cutaneous aging is a complex and cumulative process that can be subdivided into two different types: intrinsic and extrinsic. Intrinsic aging is caused by genetic and chronological factors and is similar to the aging of other organs. On the other hand, extrinsic aging is the result of exposure to environmental factors, predominantly the ultraviolet radiation emitted by solar rays [2,3,4]. Extrinsic aging of the skin, also called photoaging, is characterized by a series of physiological and structural changes that impact on the appearance of the skin. Solar elastosis is one of the processes induced by ultraviolet radiation and involves degeneration of elastin and collagen fibers with accumulation of elastotic material in the dermal layer, indicating cutaneous involvement [5, 6]. This process impacts skin elasticity, firmness, and resilience [7].
Elastin is one of the major components of the skin but is also present in a variety of vertebrate tissues and organs, such as blood vessels, cartilage, lungs, and heart [8, 9]. Together with collagen and proteoglycans, elastin makes up the extracellular matrix (ECM) that is produced by dermal fibroblasts. Elastic fibers are insoluble components essential to the ECM [10]. Numerous molecules are associated with elastic fibers, some of which are studied in this article. Fibrillins are the main structural components of microfibrillins, with fibrillin 1 being particularly important for the formation of elastic fibers [8]. Microfibril-associated glycoprotein 2 (MAPG2) is essential for cell signaling during microfibril formation and elastogenesis [11]. Lysyl oxidases (LOXs) are part of the class of molecules associated with the production of elastic fibers in which the formation of covalent bonds stabilizes the insoluble polymerized elastin product [12].
Elastosis and ECM degradation occur through enzymes capable of cleaving these molecules, including cysteine proteases, serine proteases, and matrix metalloproteinases, which contribute to the aging process [8, 12].
Elastin has a mass between 60 and 72 kDa and a polymeric structure formed from monomers of its soluble precursor, tropoelastin. Important alternative splicing occurs in the tropoelastin primary transcripts, and variations in the mRNA sequences result in the production of multiple isoforms [13]. One isoform that includes exon 26A contains a highly unusual hydrophilic domain that has not been identified in other species. This isoform is not transcribed into normal elastic cutaneous tissue and may occasionally be present in some conditions such as exposure to UV rays or extreme temperature treatments [14,15,16]. Thus, it appears that some forms of tropoelastin protein are associated with healthy elastic tissue, whereas others are associated with damage or disease [17].
When exon 26A is in the elastin transcript, 99 base pairs encoding an additional 33 amino acids (GADEGVRRSLSPELREGDPSSSQHLPSTPSSPR), compared with common tropoelastin, are added. Thus, the inclusion of exon 26A can probably alter the structural and biological properties of elastin because it includes a hydrophilic domain [14, 18, 19].
Due to the importance of elastin and elastic fibers in the composition of the dermis and its involvement in skin aging, it is necessary to better understand the biological mechanisms that result from the presence of exon 26A after photoexposure. This understanding contributes to the search for new methods to prevent and treat skin aging because reversal of the aesthetic effects of aging is being increasingly desired. Thus, the main objective of this study was to evaluate the expression of exon 26A under photoexposed conditions (acute and chronic irradiation) as well as to evaluate molecular and protein differences between elastin and other markers related to elastogenesis among different in vitro study models.
Methods
Ex Vivo Skin
Ex vivo skin from three different areas, two areas exposed to the sun (preauricular region and eyelids) and an unexposed area (breast), were obtained from a total of 73 female volunteers who were scheduled to undergo cosmetic surgery and who were healthy and showed phototypes II or III. Thirty samples from the region of the breast (control, not photoexposed), 19 from the region anterior to the ear (rhytidoplasty, photoexposed), and 24 from the region around the eyes (blepharoplasty, photoexposed) were collected. Skin samples were cut and stored in a freezer at −80 °C (Sanyo, Moriguchi, Japan) in RNA later solution (Ambion, Carlsbad, CA) until RNA extraction and subsequent analysis by qPCR. All procedures were conducted in accordance with the Declaration of Helsinki and were previously approved by the Ethics and Research Committee on Humans, under protocol no. 297.371 dated 06/07/2013. All volunteers signed the Informed Consent Form. The ages of the samples ranged from 20 to 69 years; the mean age of the control group was 46.1 ± 12.4 years, the rhytidoplasty group 54.6 ± 9.8 years and the blepharoplasty group 51.9 ± 10.4 years.
Cellular Coculture and Reconstituted Skin (3D Skin or Equivalent Skin)
A pretest was performed on cell cocultures to determine the optimal irradiation dose before conducting the reconstituted skin evaluations. Neonatal human dermal fibroblasts (HDFn) (Invitrogen, Carlsbad, CA), maintained in DMEM medium (Gibco, Carlsbad, CA) containing 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA), were used. The fibroblasts were plated in coculture with neonatal human epidermal keratinocytes (Invitrogen, Carlsbad, CA) and maintained in EpiLife medium (Gibco, Carlsbad, CA) containing human keratinocyte growth supplement (HKGS) (Invitrogen, Carlsbad, CA) and cultured at 37 °C with 6% CO2.
An irradiation dose curve (control without irradiation—1 kJ/m2—25 kJ/m2—50 kJ/m2—100 kJ/m2—200 kJ/m2—300 kJ/m2—400 kJ/m2—500 kJ/m2) was administered with a Suntest Atlas CPS + (Atlas, Mount Prospect, Illinois) solar radiation simulator. After the treatment, a viability assay using thiazolyl blue tetrazolium bromide (MTT) (Sigma-Aldrich Co., St. Louis, MO) tetrazolium salt was conducted to determine the concentration at which 80% of the cells remained alive (inhibitory concentration—IC20). Gene expression analysis of normal synthesized elastin (26–27) and altered elastin (26A) was also performed.
A total of 30 reconstituted skins were prepared, and half were subjected to the irradiation dose of 200 kJ/m2, which was previously detected in the MTT viability test, whereas the others were used as a non-photoexposed control (adapted from Fernandez et al. [46]). In vitro skin expression analysis and immunohistochemical analysis of normal elastin (Abcam, Cambridge, UK), tropoelastin (Abcam, Cambridge, UK), fibrillin 1 (FBN1) (Abcam, Cambridge, UK), LOX (Abcam, Cambridge, UK), MMP12 (Abcam, Cambridge, UK), and MAGP2 (Abcam, Cambridge, UK) were carried out on skins stored in 4% formaldehyde (Sigma-Aldrich Co., St. Louis, MO).
The reconstituted skins were prepared according to methods described by Brohem et al. [47]. Fibroblasts and keratinocytes cultured under the same conditions as the coculture cells were used. The fibroblasts were distributed on an acellular collagen matrix of the reconstituted skin at a density of 6 × 104 cells per insert and maintained in culture under the same conditions described above for 6 h until polymerization. After this period, the keratinocytes were plated on the layer containing fibroblasts at a density of 1.2 × 105 cells per well. The skins were nourished by means of reconstituted skin differentiation and maintained at the air–liquid interface for 10–13 days to promote keratinocyte differentiation and development of stratified layers, thus resulting in the formation of an epidermal layer that is histologically similar to human skin.
Histology and Immunohistochemistry
For analysis of the immunohistochemical assays, mean values of the experiments were calculated after the images were evaluated for the intensity of labeling of each antibody used. The methodology was based on the tests described by Koria et al. [48]. The morphology of the reconstituted skins was evaluated by hematoxylin and eosin staining of paraffin-embedded samples. For immunohistochemistry, the skins were fixed in 4% formaldehyde embedded in paraffin, and the blocks were subsequently deparaffinized and rehydrated by reverse ethanol/xylene washes (Sigma-Aldrich Co., St. Louis, MO). For detection of antibodies, the sections were washed with phosphate-buffered saline (PBS) (Sigma-Aldrich Co., St. Louis, MO) and incubated for 1 h at room temperature with the following primary antibodies: anti-elastin, mouse monoclonal antibody (1:200 dilution); anti-tropoelastin, rabbit polyclonal antibody (1:200 dilution); anti-fibrillin 1 (Biotin), mouse monoclonal antibody (1:25 dilution); anti-LOX, rabbit monoclonal antibody (1:75 dilution); rabbit anti-MMP12 antibody (1:50 dilution); and anti-MAGP2, rabbit monoclonal antibody (1:100 dilution). The slides were then incubated with complement reagent (Abcam, Cambridge, UK) for 10 min, followed by a biotinylated secondary antibody (Abcam, Cambridge, UK) for 15 min at room temperature. The chromogen DAB (diaminobenzidine 3,3′) (Abcam, Cambridge, UK) was added to the slides for 5 min at room temperature. The slides were then washed with PBS buffer (Sigma-Aldrich Co., St. Louis, MO). After washing, the slides were hematoxylin-countercoated and washed seven to eight times under running water before subsequent dehydration and mounting with permanent mounting medium (Abcam, Cambridge, UK).
Analysis of Gene Expression of Ex Vivo, Cocultured and Reconstituted Skins
For the extraction of RNA from ex vivo skins and reconstituted skins, the materials were previously lysed in TissueLyser (Qiagen, Venlo, Germany), and the coculture cells were detached from the plates using a trypsin–ethylenediaminetetraacetic acid (EDTA) solution (Sigma-Aldrich Co., St. Louis, MO) and then pelleted for freezing at −80 °C until extraction. The other steps were performed in the same manner for the three conditions (ex vivo skin, reconstituted skin, and cell coculture). RNA was isolated using cell lysis buffer and purified on a glass fiber column with reagents from the RNeasy Mini extraction kit (Qiagen, Venlo, Germany). The cDNA was generated from a reverse transcription reaction using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA), following the manufacturer’s recommended protocol and using a Veriti Thermal Cycler thermocycler (Applied Biosystems, Carlsbad, CA).
Gene expression was assessed by qPCR using Taqman Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA) in a ViiA 7 Real-Time PCR System (Applied Biosystems, Carlsbad, CA) thermocycler, and the plates were prepared according to the procedure recommended by the manufacturer.
Custom probes corresponding to the junction of exons 26 and 27 (Applied Biosystems, Carlsbad, CA), which was characterized as the region common to normal elastin, were used to detect the absence of exon 26A between exons 26 and 27. Probes were also synthesized for the region subjected to alternative splicing, in which the design was performed at junction 26A and 27 (Applied Biosystems, Carlsbad, CA). The designs were performed with the help of the File Builder 3.1 PC Version Software (Applied Biosystems, Carlsbad, CA). GAPDH and ACTB genes were used as constitutive expression controls. The expression was analyzed with primers for the following markers: ELN INVENT 1–2 (normal elastin probe offered by the manufacturer and validated for use), fibrillin 1 (FBN1), lysyl oxidase (LOX), the major metalloproteinase that degrades elastin (MMP12), and microfibril-associated glycoprotein 2 (MAGP2) (all from Applied Biosystems, Carlsbad, CA).
qPCR Data Analysis
PCR data interpretation and analysis were performed with viiA 7 Software V 1.1 (Applied Biosystems, Carlsbad, CA). The analysis was performed using the previously described ΔΔCT method [49, 50].
The Ct values of each sample and each first test were used. The ΔCT was calculated from the Ct values (the Ct values of the samples were subtracted from the mean Ct of the GAPDH and the ACTB genes) and averaged for each of the following conditions: breast, preauricular region, and eyelid. From this, each exposed condition was normalized in relation to the control (breast), resulting in ΔΔCT. With ΔΔCT, the fold-change value was calculated by calculating 2−ΔΔCT for the evaluation of genes differentially expressed in relation to the control.
Only modulations characterized by fold-changes ≥ 1.5 or ≤ 0.5 (representing the genes whose expression changed significantly after sun exposure) and that presented p value ≤ 0.05 and a minimum reliability of 95% were considered significant modulations of gene expression. Statistical analyses were performed using JMP Statistics Software and one-way ANOVA by multiple comparisons and Dunnett’s post-test of the exposed conditions in relation to the non-exposed controls.
Results
Effect of Irradiation on the Expression of Elastin Exon 26A during Fibroblast and Keratinocyte Coculture
The expression of normal and altered elastin isoforms was evaluated in an in vitro test system involving the coculture of fibroblasts and keratinocytes that were exposed to a radiation dose curve. According to the results of this coculture, the gene expression of normal elastin decreased at all doses tested, when compared with the unexposed control (Fig. 1A). The expression profile of the altered isoform containing elastin 26A increased from the dose of 200 kJ/m2 (Fig. 1B). Thus, the dose of 200 kJ/m2 was chosen for further testing on the reconstituted skins.
Gene expression analysis of normal and altered elastin isoforms after exposure to different doses of radiation. Fold-change values obtained from gene expression analysis of normal elastin A and altered elastin 26A B, after different doses of irradiation in cocultured fibroblasts and keratinocytes compared with non-irradiated cocultures
Gene and Protein Expression in Reconstituted Skin
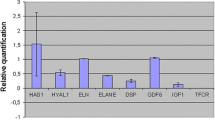
The gene expression levels of markers related to elastogenesis were determined in a reconstituted skin model. According to Fig. 2, altered elastin (ELN 26A) presented a 4.59-fold higher expression in the irradiated skins when compared with the non-irradiated samples. The MMP12 gene showed a tendency towards increased expression in irradiated skin (1.94-fold increase), but this difference was not statistically significant. Expression levels of the other markers were not significantly changed.
Evaluation of the gene expression of markers related to elastogenesis in reconstituted skin. Fold-change values obtained after analysis of gene expression of the major markers related to elastogenesis in reconstituted skins. Comparative evaluation between irradiated skins and control skins. The red line shows that values above 1 correspond to an increase in expression and those below 1 represent a decrease in expression of the analyzed genes. Statistical analysis was performed by one-way ANOVA, followed by Dunnett’s post-test *p < 0.05, **p < 0.01, ***p ≤ 0.001
The protein expression levels of markers related to elastogenesis were also evaluated in reconstituted skin with and without photoexposure. The figure below shows representative images of one experiment for each condition.
Histopathological changes were observed between the two conditions. The epidermis was thinner and atrophied with vacuolization in the cellular cytoplasm. In the dermis, the appearance of vacuolization and a decrease in the amount of fibroblasts are also visible (Fig. 3A and B).
Evaluation of protein expression through immunohistochemistry of proteins related to elastogenesis in reconstituted skin. Representative images of the samples analyzed after hematoxylin and eosin (HE) labeling (A, B) and labeling for the other markers investigated (C–N) in photoexposed and non-photoexposed skin. HE, 10× magnification. Elastin, tropoelastin, fibrillin 1, LOX, MMP12, and MAGP2, 20× magnification
Tropoelastin (Fig. 3E, F) showed a significant increase in the basal layer of the epidermis after photoexposure, and fibrillin 1 (Fig. 3G and H) was also significantly increased throughout the dermis after photoexposure. There were no changes in the protein expression levels of elastin, LOX, MMP12, and MAGP2 (Fig. 3A–D, I–N) according to this method of evaluation.
Effect of Irradiation on Gene Expression in Ex Vivo Skin
The effect of photoexposure on the gene expression of markers related to elastogenesis was also evaluated using the ex vivo skin model. According to Fig. 4, the results obtained by qPCR demonstrated increased expression of the altered elastin isoform (ELN26A) in the two exposed regions evaluated. The skin of the auricular region (rhytidoplasty) showed a 3.61-fold increase, and the skin of the eyelid region (blepharoplasty) showed a 2.58-fold increase compared with the control (non-exposed skin derived from mammoplasty). Expression of the typical elastin isoform (ELN26-27) and the elastin control (ELN INVENT 1–2) were not significantly different when compared with the expression in the non-photoexposed region. The MMP12 gene presented increased expression in the two conditions exposed, when compared with the control. The MMP12 expression by rhytidoplasty was 5.98-fold greater, and that by blepharoplasty was 3.14-fold greater compared with the control. The LOX gene, which is related to elastin crosslinking, showed increased expression of 1.64-fold only in the exposed blepharoplasty region. The FBN1 and MAGP2 genes did not show differential gene expression in the photoexposed tissues when compared with the control.
Analysis of gene expression of markers related to elastogenesis. Graph showing the fold-change values after comparison between the photoexposed areas (rhytidoplasty and blepharoplasty) in relation to the non-photoexposed area (breast). The red line indicates that values above 1 correspond to an increase in expression and those below 1 correspond a decrease in the expression of the genes analyzed in relation to the control. Rhytidoplasty includes skin samples from the anterior region to the ear, and blepharoplasty includes samples of skin around the eyes. ELN elastin, FBN1 fibrillin 1, LOX lysyl oxidase, MMP12 metalloproteinase of matrix 12, MAGP2 glycoprotein associated with microfibrils 2. Statistical analysis was performed by one-way ANOVA, followed by Dunnett’s post-test *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001
Discussion
Some researchers have reported that the presence of exon 26A may increase in aging or damaged elastic fibers and may be a marker of damage; however, the effect of this domain on the physical properties of tropoelastin and elastin is not yet known [20]. The consequences of premature aging after UV exposure are mostly related to alterations in the mechanical properties of the tissue and are the result of structural and compositional remodeling in the dermis. These processes cause wrinkling that characterizes chronically photoaged skin [21].
This study analyzed the differences in gene expression that can occur in molecules related to the production of elastic fibers in the skin after photoexposure. The main objective was to evaluate whether irradiation induces the production of a structurally altered elastin molecule (exon 26A) as well as to verify the expression of other elastic fiber-related markers due to the consequences on the skin and the signs of aging.
The evaluation of the gene expression in irradiated cells demonstrated that low doses (< 200 kJ/m2) of irradiation were insufficient to impact the expression of the altered elastin isoform (exon 26A) in a coculture model. However, at more intense radiation doses, expression of the altered elastin isoform was increased and that of the typical isoform was decreased, which indicated damage to the elastin fibers caused by extrinsic aging. Mora Huertas et al. [8] has demonstrated some molecular consequences in elastin fibers following skin aging, indicating differences in the morphological pattern and more pronounced elastin decomposition in the tissues of older patients.
The decrease in normal elastin gene expression evidenced in the studied models, ex vivo skin derived from photoexposed tissues in the reconstituted skin model and the co-culture, complement what is described in the literature because several studies have shown that the expression of normal elastin decreases after photoexposure and photoaging [14, 22,23,24]. Bernestein et al. [25] demonstrated that elastin mRNA levels decrease in photoexposed skin and in fibroblasts derived from the same skins when compared with the non-exposed areas and cells of the same individuals. Additionally, Darvin et al. [26] reported that high exposure to solar radiation significantly reduced the concentration of elastic fibers and collagen when analyzing skin material after UV exposure.
No studies in the literature have investigated the gene expression of exon 26A in cocultures of keratinocytes and fibroblasts, only in separate cell cultures of fibroblast monolayers or keratinocytes. The presence or absence of exon 26A may generate a structurally or functionally altered elastin molecule; however, the function of elastin molecules containing the additional sequence remains unknown [19]. Other studies that have evaluated specific mutations in the elastin gene have demonstrated that the charge changes caused by amino acid substitution modify the elastin structure [13, 16]. It has been confirmed that elastin is expressed by keratinocytes and that the primary elastin transcript typically contains exon 26A in the terminal differentiation of keratinocytes, demonstrating that exon 26A expression is present in skin cells [27, 28].
In vitro tests of reconstituted skin and fibroblasts were used because these are dermis cells that produce and express the elastin gene [29,30,31]. In addition, keratinocytes were used because they are cells of the epidermis that present tropoelastin expression as do fibroblasts [27, 28]. The relationship between these two cells and UV irradiation was also described by Akhalaya et al. [22] in which dermal fibroblasts were reported as the cells that were most sensitive to UV radiation. Epidermal keratinocytes were also likely involved in this process. The gene expression results obtained by the in vitro assays on reconstituted skins confirmed that exon 26A expression is indeed increased in photoexposed skins. These data correlated with ex vivo skin data, as a significant increase in the expression of the altered elastin (26A) was observed in both photoexposed ex vivo skin regions. These data also corroborated the results described by Chen et al. [14] where exon 26A mRNA expression was increased in the human epidermis in vivo when irradiated with a fluorescent lamp and after thermal shock. This work has also demonstrated the relationship between elastin 26A and solar elastosis and has assumed that elastin 26A can alter the structural and biological properties of elastin, thus affecting normal elastic fiber formation and contributing to the development of solar elastase in human skin in vivo.
In relation to the gene expression analysis of the other markers used, there was a correlation between the two methodologies tested in the in vitro and ex vivo models. Significant increases in MMP12 expression were detected in the ex vivo skin tests in both photoexposed regions, but in the irradiated reconstituted skins, this increase was not significant. MMP12 is the primary metalloproteinase that degrades elastin [32]. Chung et al. [32] have demonstrated that UV irradiation induces MMP12 gene and protein expression in human skin in vivo and in fibroblast cultures. Pandel et al. [33] demonstrated that MMPs are produced in response to UVB irradiation in vivo and are involved in changes that occur through photoaging in connective tissues. However, more recently, Tewari et al. [34] observed that the upregulation of MMP12 is more evident in UVA1 irradiation than in UVB irradiation, and thus, MMP12 would be a good marker of UVA1 exposure.
MMPs are associated with a variety of normal and pathological conditions involving matrix degradation and remodeling. Chen et al. [35] commented that solar irradiation induces the expression of MMP12 and this increased expression in photoexposed skin is colocalized with solar elastosis products. Chung et al. [32] also claimed that the protein expression of MMP12 is increased in regions of solar elastosis, suggesting that this is partly responsible for the elastic fiber damage that is universally observed in photoexposed human skins.
Fibrillin 1, a protein that is involved in the formation of elastic fibers, demonstrated diminished expression in the exposed region anterior to the ear of ex vivo skin. However, in the region around the eyes, the expression was increased when compared with the control. These differences were not significant in any of these cases. On analysis of gene expression levels in reconstituted skins, fibrillin 1 was also decreased, but these differences were not significant. In this case, only the results from the ex vivo skin of the region around the eyes corroborated the results described in the literature, i.e., Bernstein et al. [25] reported increased expression and deposition of fibrillin 1 in the reticular dermis of photodamaged skin.
The MAGP2 protein makes up the microfibrils of the extracellular matrix. It is the protein product of the MFAP5 gene and is also known as MP-25 [36]. MAGP2 colocalizes with elastin-associated microfibrils and also elastin-free microfibrils in various tissues. Lemaire et al. [37] conducted in vitro studies that suggested that the expression of MAGP2 is necessary for the production of normal elastic fibers. MAGP2 gene expression was also decreased in both in vitro and ex vivo tested conditions, but these reductions were not significant. There are still no data in the literature regarding changes in MAGP2 gene expression after photoexposure.
LOX gene expression showed a minimal decrease in the reconstituted skin and the region anterior to the ear in the ex vivo skin. On the other hand, in the region around the eyes, LOX expression was significantly increased when compared with that of the control. LOX is responsible for elastin cross-linking during elastogenesis. It has been reported that LOX expression decreases during photoaging and that this enzyme is essential for elastic fiber homeostasis and maintenance of elastogenesis [38]. Pain et al. [39] complemented that too much elastin and insufficient lysyl oxidase enzyme (LOXL1) to form functional elastic fibers may be caused by an imbalance of the elastin fibers in response to ultraviolet light. For this marker, a significant increase in gene expression was observed in the periocular region, which was not confirmed in the reconstituted skin model.
On evaluation of protein expression by immunohistochemistry method in reconstituted skins, no elastin was detected in the control and in the photoexposed conditions, and consequently, there was no modulation after photoexposure. Mewes et al. [40] studied the expression of elastin by immunohistochemistry in reconstituted skins that remained in differentiation for 50 days, but detected elastin in these skins only after 7 days of culture in an air–liquid interface. The reconstituted skins used in this study remained in differentiation for 13 days. Although the preparation of skin samples was different when compared with the Mewes methodology, this work also demonstrated that elastin labeling by immunohistochemistry does not occur in the first days after preparation of the skin samples.
On immunohistochemistry analysis of reconstituted skins, tropoelastin and fibrillin 1 protein expression levels were increased after photoexposure. LOX, MMP12, and MAGP2 showed no significant differences in expression after photoexposure. One hypothesis that may be suggested is that the method used to expose the skins to solar radiation was not effective in stimulating effects on the expression levels of the markers investigated (elastin, LOX, MMP12, and MAGP2).
Another consideration is the reconstituted skin model. This is an in vitro method that serves as a versatile and reproducible research tool with numerous applications, and is recognized and validated by the international scientific community, in addition to being indicated for substitution in animal tests on various topics such as dermal irritation. However, there are some technical limitations with respect to human skin regarding the complexity of the dermis, reduction of barrier function, absence of blood vessels, cells of the immune system, hair follicles, and glands and appendages, a fact that could justify the absence of protein labeling. Currently, due to the legal and ethical requirements regarding tests for cosmetic industries, in vitro reconstituted skin models are validated and correlated with in vivo data owing to their complexity and thus enable the construction of a predictive and robust model that can replace animal models [41].
Therefore, future studies of the protein expression levels of these markers by ex vivo skin immunohistochemistry assay should be performed, and the results should be compared with data obtained via the in vitro reconstituted skin model. For example, a study conducted by Gambichler et al. [42] analyzed another skin disease: mid-dermal elastolysis (MDE) which is a rare, acquired skin disease histopathologically characterized by selective loss of elastic fibers in the mid-dermis. In this review, they described that, to better understand pathomechanisms involved in MDE, immunohistochemistry of skin biopsies was used, and these assays proved to be a good technique for analyzed skin markers.
Complementarily, Widgerow et al. [43] studied new approaches to skin photodamage histology using a combination of stains that provided a comprehensive range of stains for identifying new elastin or regenerative events within the skin layers.
With these last considerations, we recognize that the present study has some limitations. In addition to comparing in vitro with ex vivo skin immunohistochemistry, the next step could be to carry out other complementary evaluations, for example as cited by Rathod et al. [44], who achieved quantification of desmosine and isodesmosine (cross-linking amino acids in the elastin biomolecule), using matrix-assisted laser desorption ionization (MALDI)–tandem mass spectrometry (MS2) and concluded that this technique is suitable for quantifying biomolecules.
Another alternative is Raman microscopy, as described by Egawa [45], who, due to the limitations with some conventional histological methods, cited that the latest application of coherent Raman scattering microscopy to visualize 3D intracellular morphologies in the human epidermis during differentiation is being applied to evaluate skin components. Thus, it is expected that valuable insights into the biological mechanisms involved in extrinsic skin aging (solar elastosis) could be identified and better understood.
Conclusion
In conclusion, the data obtained through gene expression analysis corroborated those reported in the literature among different study models (in vitro and ex vivo). It was observed that, although the photoaging process may be complex, it can be mitigated by avoiding skin exposure to solar radiation. Some of the biological mechanisms that are involved in photoaging can be understood with gene expression analyses of exon 26A. Although few studies on exon 26A expression are available in the literature, this study has shown that this change is actually associated with photoexposure in different in vitro models. Further research should be conducted to validate the mechanisms involved in the expression of this exon because the process by which the solar irradiation induces this change and the exact function of this altered elastin isoform remain unknown. The knowledge that elastin alteration contributes to solar elastosis, a very striking feature of extrinsic aging, can contribute to the prevention of extrinsic aging through the development of active components and specific treatment products for this purpose.
References
Gragnani A, Cornick SM, Chominski V, et al. Review of major theories of skin aging. Adv Aging Res. 2014;3(4):265–84.
Debacq-Chainiaux F, Leduc C, Verbeke A, Toussaint O. UV, stress and aging. Dermatoendocrinol. 2012;4:236–40.
Godic A, Poljšak B, Adamic M, et al. The role of antioxidants in skin cancer prevention and treatment. Oxid Med Cell Longev. 2014;2014: 860479. https://doi.org/10.1155/2014/860479.
Villaret A, Ipinazar C, Satar T, et al. Raman characterization of human skin aging. Skin Res Technol. 2019;25(3):270–6. https://doi.org/10.1111/srt.12643.
Hibbert S, Watson REB, Gibbs NK, et al. A potential role for endogenous proteins as sacrificial sunscreens and antioxidants in human tissues. Redox Biol. 2015;5:101–13.
Kvaskoff M, Pandeya N, Green AC, et al. Solar elastosis and cutaneous melanoma: a site-specific analysis. Int J Cancer. 2015;136(12):2900–11.
Naylor EC, Watson REB, Sherratt MJ. Molecular aspects of skin ageing. Maturitas. 2011;69(3):249–56.
Mora Huertas AC, Schmeizer CEH, Hoehenwarter W, et al. Molecular-level insights into aging processes of skin elastin. Biochimie. 2016;128–129:163–73.
Usuki T, Sugimura T, Komatsu A, Koseki Y. Biomimetic chichibabin pyridine synthesis of the COPD biomarkers and elastin cross-linkers isodesmosine and desmosine. Org Lett. 2014;16:1672–5.
Uehara E, Hokazono H, Hida M, et al. GABA promotes elastin synthesis and elastin fiber formation in normal human dermal fibroblasts (HDFs). Biosci Biotechnol Biochem. 2017;81(6):1198–205.
Hanssen E, Hew FH, Moore E, et al. MAGP-2 has multiple binding regions on fibrillins and has covalent periodic association with fibrillin-containing microfibrils. J Biol Chem. 2004;279(28):29185–94.
Kielty CM, Sherratt MJ, Shuttleworth AC. Elastic fibres. J Cell Sci. 2002;115(14):2817–28.
Yeo GC, Baldock C, Wise SG, et al. Targeted modulation of tropoelastin structure and assembly. ACS Biomater Sci Eng. 2017;3(11):2832–44. https://doi.org/10.1021/acsbiomaterials.6b00564.
Chen Z, Shin MH, Moon YJ, et al. Modulation of elastin exon 26A mRNA and protein expression in human skin in vivo. Exp Dermatol. 2009;18:378–86.
Weihermann AC, Lorencini M, Brohem CA, et al. Elastin structure and its involvement in skin photoageing. Int J Cosmet Sci. 2017;39(3):241–7. https://doi.org/10.1111/ics.12372.
Miao M, Reichheld SE, Muiznieks LD, et al. Single nucleotide polymorphisms and domain/splice variants modulate assembly and elastomeric properties of human elastin. Implications for tissue specificity and durability of elastic tissue. Biopolymers. 2017;107(5):1–20.
Carruthers J, Carruthers A, Alam M. Soft tissue augmentation. 3rd ed. Edinburgh: Elsevier; 2013. p. 54.
Bisaccia F, Castiglion-Morelli MA, Spisani S, et al. The amino-acid-sequence coded by the rarely expressed exon 26A of human elastin contains a stable beta-turn with chemotactic activity for monocytes. Biochemistry. 1998;37(31):11128–35.
Ostuni A, Lograno MD, Gasbarro AR, et al. Novel properties of peptides derived from the sequence coded by exon 26A of human elastin. Int J Biochem Cell Biol. 2002;34(2):130–5.
Vrhovski B, Weiss AS. Review: biochemistry of tropoelastin. Eur J Biochem. 1998;258:1218.
Watson REB, Gibbs NK, Griffiths CEM, Sherratt MJ. Damage to skin extracellular matrix induced by UV exposure. Antioxid Redox Signal. 2014;21(7):1063–77. https://doi.org/10.1089/ars.2013.5653.
Akhalaya MY, Maksimova GV, Rubina AB, et al. Molecular action mechanisms of solar infrared radiation and heat on human skin. Ageing Res Rev. 2014;16:1–11.
Amaro-Ortiz A, Yan B, Dorazio JA. Ultraviolet radiation, aging and the skin: prevention of damage by topical cAMP manipulation. Molecules. 2014;19:6202–19.
Seo JY, Lee SH, Youn CS, et al. Ultraviolet radiation increases tropoelastin mRNA expression in the epidermis of human skin in vivo. J Invest Dermatol. 2001;116:915–9.
Bernstein EF, Chen YQ, Tamai K, et al. Enhanced elastin and fibrillin gene expression in chronically photodamaged skin. J Invest Dermatol. 1994;103(2):182–6.
Darvin ME, Richter H, Ahlberg S, et al. Influence of sun exposure on the cutaneous collagen/elastin fibers and carotenoids: negative effects can be reduced by application of sunscreen. J Biophoton. 2014;7(9):735–43. https://doi.org/10.1002/jbio.201300171.
Hirano E, Okamoto K, Matsubara MT, et al. Elastin expression in cultured human keratinocytes: exon 26A of elastin primary transcript is always included in terminally differentiated keratinocytes. Arch Dermatol Re. 2001;293:430–3.
Kajiya H, Tanaka N, Inazumi T, et al. Cultured human keratinocytes express tropoelastin. J Invest Dermatol. 1997;109:641–4.
Achterberg VF, Buscemi L, Diekmann H, et al. The nano-scale mechanical properties of the extracellular matrix regulate dermal fibroblast function. J Invest Dermatol. 2014;134(7):1862–72. https://doi.org/10.1038/jid.2014.90.
Fazio MJ, Olsen DR, Kuivaniemi H, et al. Isolation and characterization of human elastic DNAs, and age-associated variation in elastin gene expression in cultured skin fibroblasts. Lab Invest. 1988;58(3):270–7.
Merrilees MJ, Falk BA, Zuo N, et al. Use of versican variant V3 and versican antisense expression to engineer cultured human skin containing increased content of insoluble elastin. J Tissue Eng Regen Med. 2017;11(1):295–305. https://doi.org/10.1002/term.1913.
Chung JH, Seo JY, Lee MK, et al. Ultraviolet modulation of human macrophage metalloelastase in human skin in vivo. J Invest Dermatol. 2002;119:507–12.
Pandel R, Poljsak B, Godic A, Dahmane R. Skin photoaging and the role of antioxidants in its prevention. ISRN Dermatol. 2013;2013: 930164. https://doi.org/10.1155/2013/930164.
Tewari A, Grys K, Kollet J, et al. Upregulation of MMP12 and its activity by UVA1 in human skin: potential implications for photoaging. J Invest Dermatol. 2014;134:2598–609.
Chen Z, Seo JY, Kim YK, et al. Heat modulation of tropoelastin, fibrillin-1, and matrix metalloproteinase-12 in human skin in vivo. J Invest Dermatol. 2005;124:70–8.
Combs MD, Knutsen RH, Broekelmann TJ, et al. Microfibril-associated glycoprotein 2 (MAGP2) loss of function has pleiotropic effects in vivo. J Biol Chem. 2013;288:28869–80.
Lemaire R, Bayle J, Mecham RP, Lafyatis R. Microfibril-associated MAGP-2 stimulates elastic fiber assembly. J Biol Chem. 2007;282:800–8.
Cenizo V, André V, Reymermier C, et al. LOXL as a target to increase the elastin content in adult skin: a dill extract induces the LOXL gene expression. Exp Dermatol. 2006;15:574–81.
Pain S, Berthélémy N, Naudin C, Degrave V, André-Frei V. Understanding solar skin elastosis—cause and treatment. J Cosmet Sci. 2018;69(3):175–85.
Mewes KR, Raus M, Bernd A, et al. Elastin expression in a newly developed full-thickness skin equivalent. Skin Pharmacol Physiol. 2007;20(2):85–95.
Mathes SH, Ruffner H, Graf-Hausner U. The use of skin models in drug development. Adv Drug Deliv Rev. 2014;69–70:81–102.
Gambichler T, Mamali K, Scheel C. A brief literature update on mid-dermal elastolysis with an emphasis on pathogenetic and therapeutic aspects. J Clin Aesthet Dermatol. 2020;13(9):E53–8.
Widgerow AD, Napekoski K. New approaches to skin photodamage histology—differentiating “good” versus “bad” Elastin. J Cosmet Dermatol. 2021;20(2):526–31. https://doi.org/10.1111/jocd.13865.
Rathod P, Kaur M, Ho HP, et al. Quantification of desmosine and isodesmosine using MALDI-ion trap tandem mass spectrometry. Anal Bioanal Chem. 2018;410(26):6881–9. https://doi.org/10.1007/s00216-018-1288-z.
Egawa M. Raman microscopy for skin evaluation. Analyst. 2021. https://doi.org/10.1039/D0AN02039G.
Fernandez TL, Lonkhuyzen DRV, Dawson RA, et al. Characterization of a human skin equivalent model to study the effects of ultraviolet b radiation on keratinocytes. Tiss Eng. 2014;20(7):588–98. https://doi.org/10.1089/ten.TEC.2013.0293.
Brohem CA, Cardeal LB, Tiago M, et al. Artificial skin in perspective: concepts and applications. Pigment Cell Melanoma Res. 2011;24(1):35–50.
Koria P, Brazeau D, Kirkwood K, et al. Gene expression profile of tissue engineered skin subjected to acute barrier disruption. J Invest Dermatol. 2003;121:368–82.
Livak KJ, Schimittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–8.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8.
Acknowledgements
The authors thank all subjects for their cooperation in the study and would like to thank the Boticário Group (Grupo Boticário) for funding the research. In memory of Dr. Márcio Lorencini, a coauthor of this work, a great researcher who promoted research in alternative methods and molecular biology in Brazil.
Funding
Funding for this study and for the journal’s Rapid Service fee was obtained from Boticário Group (Grupo Boticário), São José dos Pinhais, Paraná, Brazil.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
A.B. conceived of the presented idea, devised the project, the main conceptual ideas and proof outline, designed and performed the experiments, derived the models and analysed the data. C. contributed to the design and implementation of the research.
Disclosures
All the authors (Ana Cristina Weihermann, Camila Miranda de Carvalho, Desirée Cigaran Schuck, Bruna Bastos Swinka, Rodrigo Makowiecky Stuart, Ruth Maria Graf, Márcio Lorencini (in memory) and Carla Abdo Brohem) have no conflicts of interest to declare.
Compliance with Ethics Guidelines
All procedures were conducted in accordance with the Declaration of Helsinki and were previously approved by the Ethics and Research Committee on Humans, under Protocol No. 297.371 dated 06/07/2013. All volunteers signed the Informed Consent Form.
This study protocol was reviewed and approved by Centro Universitário Positivo – UNICENP, Plataforma Brasil, approval number 297.371.
All volunteers signed the Informed Consent Form.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Weihermann, A.C., de Carvalho, C.M., Schuck, D.C. et al. Modulation of Photoaging-Induced Cutaneous Elastin: Evaluation of Gene and Protein Expression of Markers Related to Elastogenesis Under Different Photoexposure Conditions. Dermatol Ther (Heidelb) 11, 2043–2056 (2021). https://doi.org/10.1007/s13555-021-00603-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13555-021-00603-y