Abstract
Aims
We aimed to verify the usefulness of targeted next-generation sequencing (NGS) technology for diagnosing monogenic diabetes in a single center.
Methods
We designed an amplicon-based NGS panel targeting 34 genes associated with known monogenic diabetes and performed resequencing in 56 patients with autoantibody-negative diabetes mellitus diagnosed at < 50 years who had not been highly obese. By bioinformatic analysis, we filtered significant variants based on allele frequency (< 0.005 in East Asians) and functional prediction. We estimated the pathogenicity of each variant upon considering the family history.
Results
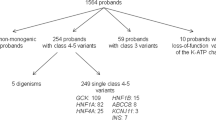
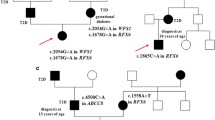
Overall, 16 candidate causative variants were identified in 16 patients. Among them, two previously known heterozygous nonsynonymous single-nucleotide variants associated with monogenic diabetes were confirmed as causative variants: one each in the GCK and WFS1 genes. The former was found in two independent diabetes-affected families. Two novel putatively deleterious heterozygous variants were also assumed to be causative from the family history: one frameshift and one nonsynonymous single-nucleotide variant in the HNF4A gene. Twelve variants remained as candidates associated with the development of diabetes.
Conclusion
Targeted NGS panel testing was useful to diagnose various forms of monogenic diabetes in combination with familial analysis, but additional ingenuity would be needed for practice.
Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Schwitzgebel VM. Many faces of monogenic diabetes. J Diabetes Investig. 2014;5(2):121–33.
Horikawa Y. Maturity-onset diabetes of the young as a model for elucidating the multifactorial origin of type 2 diabetes mellitus. J Diabetes Investig. 2018;9(4):704–12.
Alkorta-Aranburu G, Carmody D, Cheng YW, Nelakuditi V, Ma L, Dickens JT, Das S, Greeley SAW, Del Gaudio D. Phenotypic heterogeneity in monogenic diabetes: the clinical and diagnostic utility of a gene panel-based next-generation sequencing approach. Mol Genet Metab. 2014;113(4):315–20.
Ang SF, Lim SC, Tan CSh, Fong JC, Kon WY, Lian JX, Subramanium T, Sum CF. A preliminary study to evaluate the strategy of combining clinical criteria and next generation sequencing (NGS) for the identification of monogenic diabetes among multi-ethnic Asians. Diabetes Res Clin Pract. 2016;119:13–22.
Ushijima K, Fukami M, Ayabe T, Narumi S, Okuno M, Nakamura A, Takahashi T, Ihara K, Ohkubo K, Tachikawa E, Nakayama S, Arai J, Kikuchi N, Kikuchi T, Kawamura T, Urakami T, Hata K, Nakabayashi K, Matsubara Y, Amemiya S, Ogata T, Yokota I, Sugihara S, Japanese Study Group of Insulin Therapy for Childhood and Adolescent Diabetes. Comprehensive screening for monogenic diabetes in 89 Japanese children with insulin-requiring antibody-negative type 1 diabetes. Pediatr Diabetes. 2018;19(2):243–50.
Donath X, Saint-Martin C, Dubois-Laforgue D, Rajasingham R, Mifsud F, Ciangura C, Timsit J, Bellanné-Chantelot C, Monogenic Diabetes Study Group of the Société Francophone du Diabète. Next-generation sequencing identifies monogenic diabetes in 16% of patients with late adolescence/adult-onset diabetes selected on a clinical basis: a cross-sectional analysis. BMC Med. 2019;17:132.
Tatsi EB, Kanaka-Gantenbein C, Scorilas A, Chrousos GP, Sertedaki A. Next generation sequencing targeted gene panel in Greek MODY patients increases diagnostic accuracy. Pediatr Diabetes. 2020;21(1):28–39.
Ivanoshchuk DE, Shakhtshneider EV, Rymar OD, Ovsyannikova AK, Mikhailova SV, Fishman VS, Valeev ES, Orlov PS, Voevoda MI. The mutation spectrum of maturity onset diabetes of the young (MODY)-associated genes among western siberia patients. J Pers Med. 2021;11(1):57.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–24.
Yorifuji T, Fujimaru R, Hosokawa Y, Tamagawa N, Shiozaki M, Aizu K, Jinno K, Maruo Y, Nagasaka H, Tajima T, Kobayashi K, Urakami T. Comprehensive molecular analysis of Japanese patients with pediatric-onset MODY-type diabetes mellitus. Pediatr Diabetes. 2012;13(1):26–32.
Khanim F, Kirk J, Latif F, Barrett TG. WFS1/wolframin mutations, wolfram syndrome, and associated diseases. Hum Mutat. 2001;17(5):357–67.
Matsunaga K, Tanabe K, Inoue H, Okuya S, Ohta Y, Akiyama M, Taguchi A, Kora Y, Okayama N, Yamada Y, Wada Y, Amemiya S, Sugihara S, Nakao Y, Oka Y, Tanizawa Y. Wolfram syndrome in the Japanese population; molecular analysis of WFS1 gene and characterization of clinical features. PLoS ONE. 2014;9(9): e106906.
Gómez-Zaera M, Strom TM, Rodríguez B, Estivill X, Meitinger T, Nunes V. Presence of a major WFS1 mutation in Spanish Wolfram syndrome pedigrees. Mol Genet Metab. 2001;72(1):72–81.
Aloi C, Salina A, Pasquali L, Lugani F, Perri K, Russo C, Tallone R, Ghiggeri GM, Lorini R, d’Annunzio G. Wolfram syndrome: new mutations, different phenotype. PLoS ONE. 2012;7(1): e29150.
American Diabetes Association Professional Practice Committee. 2 classification and diagnosis of diabetes: standards of medical care in diabetes-2022. Diabetes Care. 2022;45(Suppl 1):S17–38.
Horikawa Y, Enya M, Fushimi N, Fushimi Y, Takeda J. Screening of diabetes of youth for hepatocyte nuclear factor 1 mutations: clinical phenotype of HNF1β-related maturity-onset diabetes of the young and HNF1α-related maturity-onset diabetes of the young in Japanese. Diabet Med. 2014;31(6):721–7.
Horikawa Y, Enya M, Mabe H, Fukushima K, Takubo N, Ohashi M, Ikeda F, Hashimoto KI, Watada H, Takeda J. NEUROD1-deficient diabetes (MODY6): identification of the first cases in Japanese and the clinical features. Pediatr Diabetes. 2018;19(2):236–42.
Uraki S, Furuta H, Miyawaki M, Matsutani N, Shima Y, Iwamoto M, Matsuno S, Morita S, Furuta M, Doi A, Iwakura H, Ariyasu H, Nishi M, Suzuki H, Akamizu T. Neonatal diabetes caused by the heterozygous pro1198leu mutation in the ABCC8 gene in a male infant: 6-year clinical course. J Diabetes Investig. 2020;11(2):502–5.
Takeuchi T, Ishigaki Y, Hirota Y, Hasegawa Y, Yorifuji T, Kadowaki H, Akamizu T, Ogawa W, Katagiri H. Clinical characteristics of insulin resistance syndromes: a nationwide survey in Japan. J Diabetes Investig. 2020;11(3):603–16.
De Franco E, Shaw-Smith C, Flanagan SE, Shepherd MH, Hattersley AT, Ellard S, International NDM Consortium. GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes. 2013;62(3):993–7.
Velho G, Blanché H, Vaxillaire M, Bellanné-Chantelot C, Pardini VC, Timsit J, Passa P, Deschamps I, Robert JJ, Weber IT, Marotta D, Pilkis SJ, Lipkind GM, Bell GI, Froguel P. Identification of 14 new glucokinase mutations and description of the clinical profile of 42 MODY-2 families. Diabetologia. 1997;40:217–24.
Chakera AJ, Steele AM, Gloyn AL, Shepherd MH, Shields B, Ellard S, Hattersley AT. Recognition and management of individuals with hyperglycemia because of a heterozygous glucokinase mutation. Diabetes Care. 2015;38(7):1383–92.
Acknowledgements
The authors thank the patients and their family members who participated in this study. The authors are also grateful to Ms. Tomomi Seino, Mr. Yuki Miyano, and Ms. Ryoko Murakami for technical assistance. The authors gratefully acknowledge Dr. Takashi Daitsu of Mamigasaki Children’s Clinic, Yamagata, Japan for providing us with additional information and helpful discussions about the patients. Finally, the authors also thank Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. The idea of the study was conceived by SS. Data collection was performed by YH, KN, NT, SK, WK, and CN. Data analysis was performed by HS. The first draft of the manuscript was written by KT and all authors commented on previous versions of the manuscript. KI supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human rights statement
Name of institutional or national ethical committee on human experimentation. The Ethical Review Committee of Yamagata University Faculty of Medicine.
Date of approval
June 6th, 2017.
Approval number
2017–108
Informed consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and/or with the Helsinki Declaration of 1964 and later versions. Informed consent or substitute for it was obtained from all patients for being included in the study. For patients between the ages of 5 and 18, written informed consent was obtained from each patient or their parents/legal guardians and informed assent was given for themselves.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Takase, K., Susa, S., Sato, H. et al. Identification of causative gene variants for patients with known monogenic diabetes using a targeted next-generation sequencing panel in a single-center study. Diabetol Int 15, 203–211 (2024). https://doi.org/10.1007/s13340-023-00669-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13340-023-00669-3