Abstract
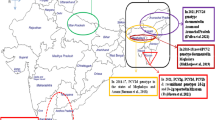
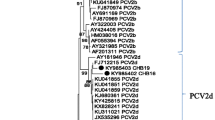
The present study examined 434 field samples including serum (n = 273), swabs from natural orifices (n = 52) and postmortem tissue samples (n = 109) from both suspected and asymptomatic swine from Andhra Pradesh, Karnataka, Kerala, Pondicherry, Tamil Nadu, and Telangana states in southern India. All the samples were processed for molecular screening of PCV3 by specific PCR assay. Overall molecular positivity rate of PCV3 was found to be 0.7% in southern India with one sample positive from each state of Tamil Nadu, Kerala and Telangana. All the three PCR positive PCV3 samples are detected from reproductive failures and were processed and propagated in PK15 cell line for virus isolation. Out of 3 samples processed, one (INDKL9PK76) PCV3 isolate could be obtained in this study and it was confirmed by specific PCR at third and fifth passage levels. Sequencing of PCV3 positive PCR amplicon (INDKL9PK76) revealed 1004 nucleotides and BLAST analysis confirmed partial sequence of the PCV3 genome. The aligned contig sequence was submitted to GenBank under the accession number of MW627201. PCV3 sequence in this study revealed 99% homology with PCV3 isolates from Europe and China. Phylogentic analysis of the PCV3 isolate-INDKL9PK76 sequence along with established PCV3 genotypes revealed clustering within PCV3 genotypes. Characterization of PCV3 (INDKL9PK76) isolate based on deduced amino acid composition of PCV3-capsid protein revealed “A” (alanine) and “R” (arginine) at 24th and 27th residues respectively confirming the incidence of PCV3a genotype. This study evidences PCV3 associated reproductive failure in domestic pigs for the first time in southern India.




Similar content being viewed by others
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Abbreviations
- PCV3:
-
Porcine circovirus 3
- ORF2:
-
Open reading frame
- MEGA:
-
Molecular evolutionary genetics analysis
- PDNS:
-
Porcine dermatitis and nephropathy syndrome
- NCBI:
-
National Center for Biotechnology Information
- BLAST:
-
Basic local alignment search tool
- PCR:
-
Polymerase chain reaction
- MLT:
-
Maximum likelihood method
- NJ:
-
Neighbor-joining
References
Bera BC, Choudhary M, Anand T, Virmani N, Sundaram K, Choudhary B, Tripathi BN. Detection and genetic characterization of porcine circovirus 3 (PCV3) in pigs in India. Transbound Emerg Dis. 2019;67(3):1062–7. https://doi.org/10.1111/tbed.13463.
Chang CC, Wu CW, Chang YC, Wu CY, Chien MS, Huang C. Detection and phylogenetic analysis of porcine cirvovirus type 3 in Taiwan. Arch Virol. 2021;166:259–63.
Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Invest. 1999;11:3–14.
Franzo G, Legnardi M, Hjulsager CK, Klaumann F, Larsen LE, Segales J, Drigo M. Full-genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within-Europe genetic heterogeneity. Transbound Emerg Dis. 2018;65(3):602–6.
Franzo G, Legnardi M, Tucciarone CM, Drigo M, Klaumann F, Sohrmann M, Segalés J. Porcine circovirus type 3: a threat to the pig industry? Vet Rec. 2018;182(3):83. https://doi.org/10.1136/vr.k91.
Franzo G, Tucciarone CM, Drigo M, Cecchinato M, Martini M, Mondin A, Menandro ML. First report of wild boar susceptibility to porcine circovirus type 3: High prevalence in the colli euganei regional park (Italy) in the absence of clinical signs. Transbound Emerg Dis. 2018;65(4):957–62.
Fu X, Fang B, Ma J, Liu Y, Bu D, Zhou P, Wang H, Jia K, Zhang G. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound Emerg Dis. 2018;65:e296–303.
Gu W, Shi Q, Wang L, Zhang J, Yuan G, Chen S, Fan J. Detection and phylogenetic analysis of porcine circovirus 3 in part of northern China from 2016 to 2018. Arch Virol. 2020;165(9):2003–11.
Karuppannan AK, Ramesh A, Reddy YK, Ramesh S, Mahaprabhu R, Jaisree S, Roy P, Sridhar R, Pazhanivel N, Sakthivelan SM, Sreekumar C, Murugan M, Jaishankar S, Gopi H, Purushothaman V, Kumanan K, Babu M. Emergence of porcine circovirus 2 associated reproductive failure in Southern India. Transbound Emerg Dis. 2016;63(3):314–20. https://doi.org/10.1111/tbed.12276.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. https://doi.org/10.1093/molbev/msy096.
Mora-Díaz J, Piñeyro P, Shen H, Schwartz K, Vannucci F, Li G, Arruda B, Gimenez-Lirola L. Isolation of PCV3 from perinatal and reproductive cases of PCV3-associated disease and in vivo characterization of PCV3 replication in CD/CD growing pigs. Viruses. 2020. https://doi.org/10.3390/v12020219.
Nguyen VG, Chung HC, Huynh TML. Molecular characterization of novel porcine circovirus 3 (PCV3) in pig populations in the North of Vietnam. Arch Gene Genome Res. 2018;1(1):24–32.
Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. Virol J. 2016;91(1):e01879-e1916. https://doi.org/10.1128/JVI.01879-16.
Parthiban S, Ramesh A, Karuppannan AK, Dhinakar Raj G, Johnson Rajeswar J, Hemalatha S, Jaisree S, Senthilkumar K, Bakasubramanyam D, Parthiban M, Sarathchandra G. Emergence of novel Porcine circovirus 2 genotypes in Southern India. Transbound Emerg Dis. 2021. https://doi.org/10.1111/tbed.14158.
Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a Circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016;13(1):184. https://doi.org/10.1186/s12985-016-0642-z.
Qi S, Su M, Guo D, Li C, Wei S, Feng L, Un D. Molecular detection and phylogenetic analysis of porcine Circovirus type 3 in 21 Provinces of China during 2015–2017. Transbound Emerg Dis. 2019;66(2):1004–15. https://doi.org/10.1111/tbed.13125.
Ren L, Chen X, Ouyang H. Interactions of porcine circovirus 2 with its hosts. Virus Genes. 2016;52(4):437–44.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Saporiti V, Martorell S, Cruz TF, Klaumann F, Correa-Fiz F, Balasch M, Sibila M, Segalés J. Frequency of detection and phylogenetic analysis of porcine circovirus3 (PCV-3) in healthy primiparous and multiparous sows and their mummified fetuses and stillborn. Pathogens (Basel, Switzerland). 2020;9(7):533. https://doi.org/10.3390/pathogens9070533.
Shulman LM, Davidson I. Viruses with circular single-stranded DNA genomes are everywhere! Annu Rev Virol. 2017;4(1):159–80. https://doi.org/10.1146/annurev-virology-101416-041953.
Tan CY, Lin C, Ooi PT. What do we know about porcine circovirus 3 (PCV3) diagnosis so far?: a review. Transbound Emerg Dis. 2021. https://doi.org/10.1111/tbed.14185.
Tan CY, Opaskornkul K, Thanawongnuwech R, Arshad SS, Hassan L, Ooi PT. First molecular detection and complete sequence analysis of porcine circovirus type 3 (PCV3) in Peninsular Malaysia. PLoS ONE. 2020;15(7): e0235832.
Vargas-Bermudez DS, Campos FS, Bonil L, Mogollon D, Jaime J. First detection of porcine circovirus type 3 in Colombia and the complete genome sequence demonstrates the circulation of PCV3a1 and PCV3a2. Vet Med Sci. 2019;5(2):182–8. https://doi.org/10.1002/vms3.155.
Wang D, Mai J, Lei B, Zhang Y, Yang Y, Wang N. Structure, antigenic properties, and highly efficient assembly of PCV4 capsid protein. Front Vet Sci. 2021;8: 695466. https://doi.org/10.3389/fvets.2021.695466.
Yang K, Jiao Z, Zhou D, Guo R, Duan Z, Tian Y. Development of a multiplex PCR to detect and discriminate porcine circoviruses in clinical specimens. BMC Infect Dis. 2019;19(1):778.
Yue W, Liu Y, Zhang X, Ma H, He J. Molecular detection of porcine circovirus type 3 in Shanxi Province, China. Anim Dis. 2021;1:8. https://doi.org/10.1186/s44149-021-00008-6.
Acknowledgements
The authors are grateful to the Professor and Head, Department of Veterinary Microbiology, MVC, Chennai-7, Professor and Head, Department of Veterinary Microbiology, Veterinary college and Research Institute, Tirunelveli, Professor and Head, Vaccine Research Centre-Viral vaccine, Centre for animal Health Studies, TANUVAS, Chennai-51 and Tamil Nadu Veterinary and Animal Sciences University (TANUVAS), Chennai-51, India for giving the necessary funds, facilities and approval to carry out this research work.
Funding
Nil.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Parthiban, S., Ramesh, A., Dhinakar Raj, G. et al. Molecular evidence of porcine circovirus 3 infection in swine: first report in southern India. VirusDis. 33, 284–290 (2022). https://doi.org/10.1007/s13337-022-00778-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-022-00778-8